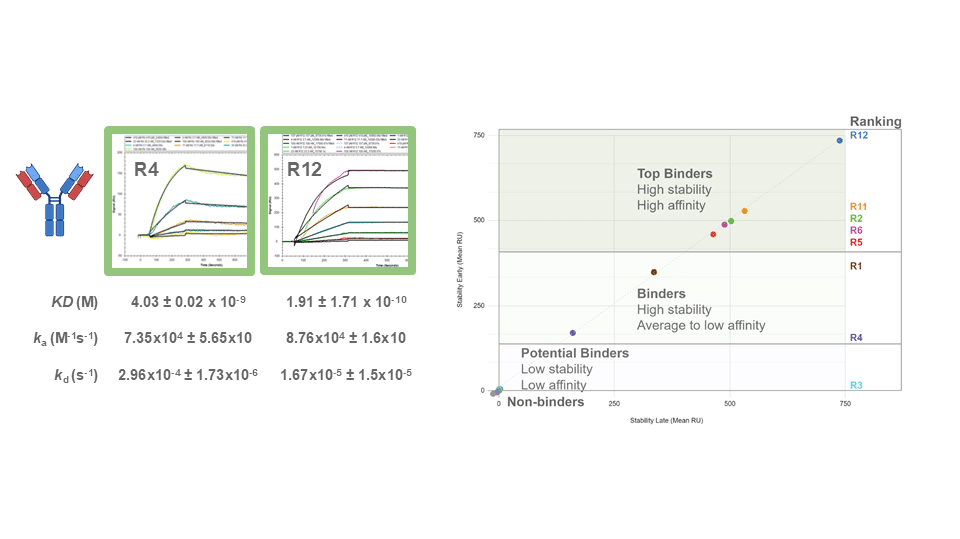
Our Technology.
Our services are built on our proprietary software for Next Generation Protein Sequencing and patented chemistry. Read on to learn about how it works and our ongoing advances in antibody sequencing, discovery and characterization technology.
- Introduction to De Novo Protein Sequencing – Webinar
- Isoleucine and Leucine: How Are They Distinguished with Mass Spectrometry? – Article
- DNA Sequencing vs Next Generation Protein Sequencing: How Do They Compare? – Article
- What is Polyclonal Antibody Sequencing? – Article
- Technologies for Antibody Discovery and Generation – Article
- Polyclonal Antibody Sequencing with Only Proteomics – Webinar
- De Novo Protein Sequencing of Antibodies for Identification of Neutralizing Antibodies in Human Plasma Post SARS-CoV-2 Vaccination – Nature Communications Article
We encourage you to dive into more of our resources below. Contact us for more information.
Live Webinar: July 22, 2025 | 11:00AM EST
Anti-Idiotype: Test Then Express
Functional Pre-Selection Accelerates Antibody Discovery
Fill out the form below to register for the webinar.
Latest Posts
Resources
Key Pain Points in Amino Acid Sequencing & How to Avoid Them
Amino acid sequencing is commonly performed using Edman degradation or mass spectrometry (MS). While mass spectrometry is favoured for its high throughput capabilities and ease of use, both techniques possess their own features and limitations. This article summarizes some of the key pain points inherent in the two methodologies when determining the amino acid sequence.
De Novo Protein Sequencing vs DNA Sequencing
DNA sequencing is the process of determining the precise order of four nucleotides bases—adenine (A), guanine (G), cytosine (C), and thymine (T)—that make up the DNA molecule. From Sanger sequencing to next-generation sequencing (NGS), DNA sequencing’s accessibility and ease of use make it one of the most widely used technologies in life sciences.
How to Determine Peptide Sequences
Amino acids (aa)—the building blocks of proteins—are simple molecules characterized by a variable R group flanked either side by an amino group and a carboxyl group. With around 20 different commonly found amino acids, each one can bond with another to produce chains that can be classified as peptides (typically below 50 aa) and proteins (sequences above 50 aa)—molecules ubiquitous to every known organism.
What is Protein Sequencing by Mass Spectrometry?
Nowadays, DNA sequencing is so popular that it is easy to forget that the first sequenced biological material was protein – insulin, by Sanger. Sanger, and another researcher, Edman, separately pioneered protein sequencing.
Antibody Protein Sequence Analysis Using Mass Spectrometry
One of the most important pieces of information researchers need to know during early stage antibody drug research and development is the sequence information of the antibody protein. With the advancement of mass spectrometry instrumentation and technologies, it is helpful, and sometimes critical, to conduct sequence analysis using mass spectrometry experiments.
Isoleucine and Leucine
Because they share the same mass, isoleucine and leucine are known as isobaric amino acids. Conventional mass spectrometry-based proteomics cannot be easily used to distinguish between isoleucine and leucine.
Ushering the New Era in Anti-Drug Antibody Assays with Next Generation Protein Sequencing
Anti-drug antibody (ADA) assays are critical to assess the clinical efficacy and safety of a biological drug and rely on control reagents that mimic the ADA response to the biological drug being tested. These positive controls typically consist of animal-derived pooled polyclonal antibodies or human monoclonal antibody reference panels against the target protein drug.
DNA Sequencing vs Next Generation Protein Sequencing
The protein sequence is key to understanding the function of a protein target and is critical to therapeutic and diagnostic development. This is particularly important for antibodies whose code diversity and glycosylation impact both function, and stability.
Enhanced Antibody Engineering for Development of Therapeutic Antibodies
Mouse monoclonal antibodies (mAbs) are highly attractive for manipulation for therapeutic applications as their manufacturing is relatively easy and well-established compared to mAbs derived from larger animal models. However, they also pose several challenges which limit their use as therapeutic agents.
The Underlying Cause of Medical Diagnostic Invalidation
In-vitro diagnostics (IVDs) are one of the most commonly used tools to diagnose conditions and guide treatment decisions and are often considered the “silent champion” of healthcare. They work by detecting the absence or presence of particular markers or by measuring the concentration of analytes or specific substances.
Increased De Novo Protein Sequencing Coverage with Optimal Protease Cocktail
In this study, we conducted a large-scale statistical analysis of protein sequencing data from samples digested with multiple proteases to understand the impact of using different combinations of proteases to improve the depth of sequence coverage in the application of de novo protein sequencing.
A Large-Scale Comparison of MS-based Antibody De Novo Protein Sequencing and Targeted DNA Sequencing
The DNA sequences of antibodies are highly diverse due to the V-(D)-J recombination and hypersomatic mutations. As such, relying on homology-based searches to sequence novel antibodies can introduce bias to sequences obtained from proteomics approaches.
In-Depth Characterization of Monoclonal Antibodies with a Single Experiment and Fully Automated Data Analysis
To develop robust mAb biologics, it is vital to fully characterize the protein, including its primary sequence, mutations, and important post-translational modifications
Recombinant Antibodies: A New Generation Enabled by Protein Sequencing
Recombinant antibodies are artificially synthesized antibodies. Recombinant antibodies are generated from expression systems (e.g., E.coli, yeast, mammalian cell lines) via transfection with two separate plasmids encoding the amino acid sequences for the light and heavy chains, respectively. In order to recombinantly produce mAbs, the amino acid sequence of the light and heavy chains must be known. There are many ways to obtain the sequence of an antibody.
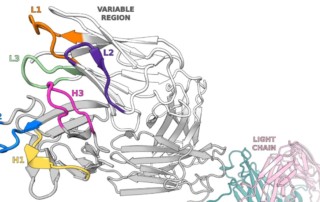
Identifying CDRs by Antibody Sequencing
The acronym “CDR” stands for complementarity determining region, a variable sequence of amino acids that folds into loops capable of binding to an antigenic amino acid sequence, also known as an epitope
Introduction to De Novo Protein Sequencing
In this on-demand webinar, our scientific sales executive Jennifer, will briefly cover the fundamentals of protein sequencing, how researchers have benefited from implementing protein sequencing into their pipelines, and discuss how Rapid Novor is able to routinely and robustly achieve 100% accuracy and 100% coverage for both monoclonal and oligoclonal antibodies.
Leveraging Recombinant Patient Antibodies in Therapeutic Applications
Our team has perfected the art of monoclonal antibody sequencing and is now ready to demonstrate our ability to sequence mAbs from polyclonal mixtures. In this talk, Anthony will walk through our new polyclonal sequencing platform that uses both proteomics and genomics to sequence the most abundant antibodies found in polyclonal sera.
Research Insurance with De Novo Protein Sequencing
If you could have guaranteed stability, certainty, and reproducibility for your research, would you be interested? Imagine this, if you’re 2 years into your project and your freezer died along with all of your important cell lines, what would you do? This is just one of the situations covered in this webinar, along with many other solutions researchers have begun to implement to safeguard their efforts. Whether you’re looking to proceed with stability and certainty or you’re looking for an immediate solution for your current reproducibility challenges, protein sequencing may be the answer.
Antibody Sequencing as a Tool to Improve Reproducibility in the Life Sciences
At PepTalk 2021, we discussed the importance of antibody standardization and explained why it’s crucial for the longevity of your research. You can listen to the full on-demand video here.
Leveraging Polyclonal Antibody Sequencing to Get High Affinity Binders Against Tough Targets
When it comes to polyclonal antibodies, how they are discovered can be just as important as how they are reproduced. In our talk originally presented at Antibody Engineering & Therapeutics Digital 2021, we highlighted the latest technology that’s capable of capturing the most abundant and high-affinity monoclonal antibodies directly from a poly mixture.