Written by:
Written by:
Vanessa Yoon Calvelo, PhD
Updated: January 18, 2023
(Published: September 15, 2022)
Analyzing Biomolecular Interactions is Important to Understanding Biological Systems
Biological processes are driven by molecules that interact through specific molecular contacts, often to form a stable complex. These interactions are typically defined by the principles of thermodynamics as well as biomolecular structure and recognition. At the simplest level is the interaction between a target molecule with a specific binding site and a probing molecule that binds to that site, resulting in the bound complex. Probing molecules can vary from small molecules to large protein complexes, which can bind to their target of interest via highly specific (ie. binding to a single site) or nonspecific (ie. binding to similar sites in a target class) interactions. Each year, new protein and DNA sequences and structures are submitted to databases, such as UNIPROT and GenBank, which only increases our need for efficient and accurate methods to characterize the myriad of biomolecular interactions that occur in any given biological system.
Analytical Techniques for Biomolecular Interactions
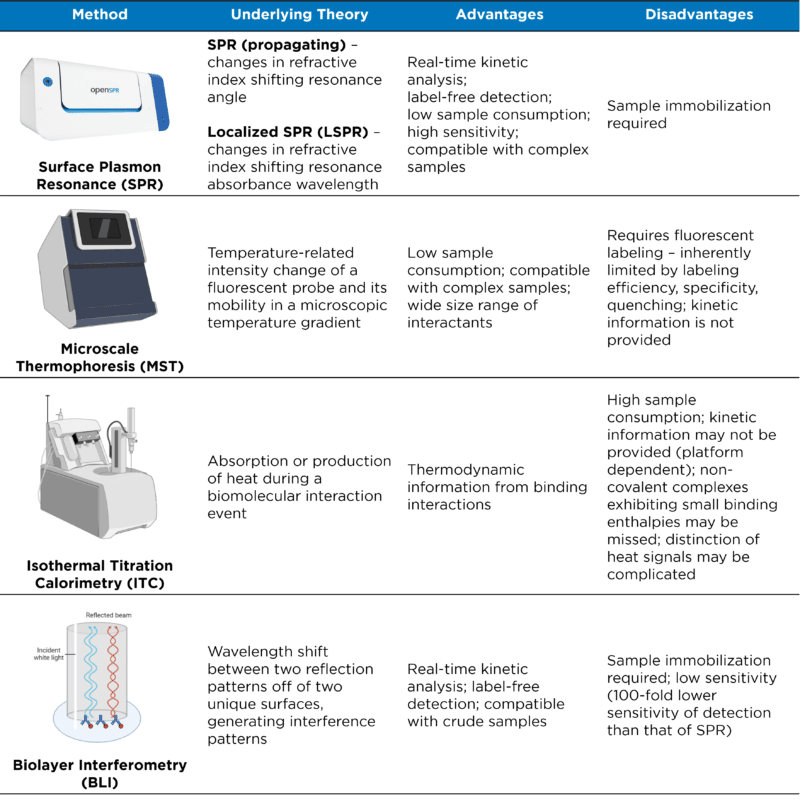
Many techniques, including surface plasmon resonance (SPR), microscale thermophoresis (MST), isothermal titration calorimetry (ITC), and biolayer interferometry (BLI), enable the characterization of biomolecular interactions, providing metrics on binding affinities, kinetics, and/or thermodynamics (Table 1). Binding affinity is often used to determine the strength of an interaction between two biomolecules; however, binding affinity is only one part of understanding the nature of the biomolecular interaction. The addition of binding kinetics (e.g. the speed of binding and release) can help achieve a fuller understanding through the characterization of unique kinetic parameters that result in the affinity observed at equilibrium (KD).
SPR is one of the most used analytical techniques for biomolecular interactions due to its versatility. MST and ITC lack the capacity to provide real-time binding affinity and kinetic measurements, while SPR can provide this information with greater accuracy and sensitivity than BLI. Therefore, SPR is widely considered a choice method in the analysis of biomolecular interactions across many applications, as discussed below.
For an overview of the SPR method and underlying theory, please refer to our Surface Plasmon Resonance Spectroscopy article.
Table 1. Common analytical techniques for characterizing biomolecular interactions.
SPR Applications for Biomolecular Interactions
The versatility of SPR is observed in its application to the characterization of diverse biomolecules and their interactions (Figure 1). In this section, we will delve further into the utility of SPR for binding kinetic analysis of some of the most fundamental biomolecular interactions.
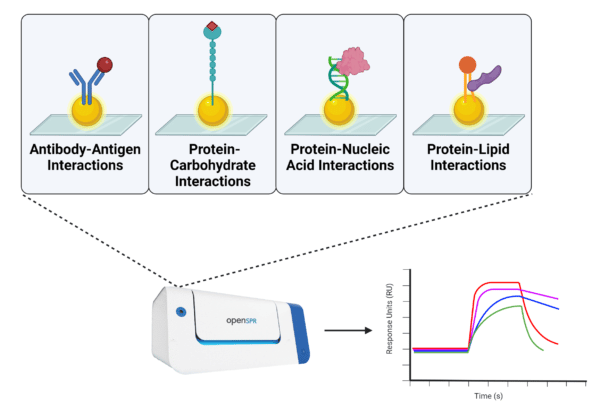
Figure 1. Examples of different types of biomolecules and their interactions characterized by SPR.
Antibody-Antigen Interactions
Monoclonal antibodies (mAbs) are widely used as diagnostic and therapeutic products in various applications, including food analysis, veterinary medicine, drug development, and biomedical research. Therefore, it is essential to understand their fundamental properties including their epitopes, kinetics, specificity, and affinity. Currently, there are several analytical techniques that can be used to determine some of these parameters, but none can evaluate all of them efficiently and accurately. Immunological techniques, such as enzyme-linked immunosorbent assay (ELISA) and Western blotting may be routinely used; however, these endpoint assays cannot provide the stoichiometric, kinetic, and/or thermodynamic information for the resulting immunocomplex.
In this regard, SPR has been successfully employed as a standard, orthogonal technique in the characterization of antibodies and their interactions. Some examples in the literature include:
- The application of SPR, immunoprecipitation, and Western blotting in the evaluation of epratuzumab, a humanized mAb that binds to the CD22 glycoprotein of mature and malignant B cells
- The application of SPR and matrix-assisted laser desorption/ionization mass spectrometry (MALDI MS) in the characterization of 1E5, a mAb that recognizes and interacts with the bovine prion protein
- The application of SPR, along with chaotrope-based ELISAs, in the characterization of antibody avidity against the immunodominant Mycobacterium tuberculosis antigen, Ag85A
Protein-Carbohydrate Interactions
Carbohydrates and their conjugates with proteins and lipids play crucial roles in cellular processes ranging from cell communication to cell death. For example, glycoproteins have been linked to numerous processes including cell adhesion, molecular trafficking, receptor activation, protein folding, and protein degradation. Aberrant glycosylation events are often implicated in pathological phenotypes (ie. cancer, diabetes, neurological disorders, immunological disorders) making these glycoforms, and the processes they are associated with, important targets for analysis and monitoring.
Structural elucidation of carbohydrates and glycoforms is inherently challenging due to their structural diversity from molecule to molecule, often necessitating sample preparation and manipulation prior to analysis. Conventional techniques, such as carbohydrate microarrays, chromatography, electrophoresis, and mass spectrometry, are sensitive but require purification, digestion, and/or enrichment steps. Furthermore, radioactive or fluorescent labels may be required for the detection and visualization of these eluted molecules.
Label-free analytical techniques have become more appealing in the characterization of carbohydrate-containing molecules. Among these techniques, SPR receives specific attention for its ability to provide real-time information on the binding affinity and kinetics of carbohydrate-specific interactions. One case study demonstrated the sensitivity of SPR in the analysis and screening of a lectin panel against serum glycoproteins. The lectin panel displayed a selective glycan recognition pattern, increasing the capacity to distinguish glycoproteins that have similar glycan structures. Another case study involved the application of SPR, as an orthogonal method, in characterizing the conformational epitope of the type III group B streptococcal polysaccharide (GBSPIII) from Streptococcus agalactiae. Two GBSPIII-specific mAbs were analyzed, using SPR and ELISA inhibition, against a large panel of oligosaccharide probes to confirm the minimum epitope binding unit and chain length for optimal interaction.
Protein-Nucleic Acid Interactions
Nucleic acids are involved in numerous cellular processes from storage, transmission, and expression of genetic information to regulation. The advent of genome sequencing has made it feasible to elucidate the links between DNA and numerous biological as well as pathological phenotypes. With the identification of other nucleic acid families, such as small interfering RNAs (siRNAs) and microRNAs (miRNAs), our understanding of diverse health and disease conditions has only advanced further.
As one of the most fundamental biomolecules of life, nucleic acids have been the target of many studies employing various bioanalytical techniques. SPR is among these techniques as a more robust, highly sensitive, label-free option. Short oligonucleotides to PCR products to larger molecules, such as cDNA and 16S ribosomal RNA, have all been subject to characterization by SPR. Of particular interest is the binding kinetic information obtained from the interactions between proteins and nucleic acids, as demonstrated in the case study of the regulator of calcineurin 1 (RCAN1) protein. RCAN1 is a multifunctional protein involved in neurodegeneration, mitochondrial dysfunction, inflammation, and protein glycosylation, and is linked to the pathogenesis of Down Syndrome. This recent case study indicates that RCAN1 is a novel RNA-binding protein that binds to a 23-nucleotide sequence of the adenine nucleotide translocator (ANT1) mRNA. The application of various analytical techniques, including RNA immunoprecipitation-sequencing (RIP-seq) and SPR, revealed the RNA-binding characteristics of RCAN1, which have also led to the discovery of an RNA aptamer that inhibits RCAN1 and provides a neuroprotective role.
Protein-Lipid Interactions
SPR is also a useful technique in the characterization of lipid-protein interactions, more specifically the interactions between proteins and the lipid bilayer of the cell membrane. The lipid bilayer is a highly polarized, dynamic structure that is composed of >1000 different lipids, including glycerophospholipids, sphingolipids, and sterols. This dynamic variety contributes to the spatial and temporal architecture of the membrane, providing a structural layout that is optimal for interacting with peripheral proteins (ie. pore-forming proteins, coagulation factors, enzymes, amyloidogenic proteins). These high-affinity interactions often regulate lipid signaling and trafficking events from cellular membranes, thus making them important targets for analysis. SPR is among the well-established biochemical and bioanalytical methods for accurately measuring lipid specificity and membrane affinity of many peripheral proteins. Examples include the binding of the phosphatidylinositol-specific phospholipase C-δ (PLC- δ) isoform (a membrane enzyme that cleaves phospholipids) and the binding of coagulation factor Va to phospholipid membranes.
SPR Assay – A golden standard for biomolecular interaction analysis
The clear advantages to SPR are such that it can provide real-time, label-free, continuous monitoring and detection of diverse biomolecular interactions, as highlighted above. Though its applications are far-reaching, assay development and optimization (ie. immobilization procedure, selection of assay buffer, choice in positive and negative controls, evaluation criteria) are often required and need to be worked out case by case, as with any other bioanalytical technique. Nonetheless, SPR is considered the golden standard for providing optimal, quality binding kinetic information for a wide variety of ligands and analytes.
Rapid Kinetic Analysis With SPR Biacore
Here at Rapid Novor, we employ a nanotechnology-based localized SPR platform to provide comprehensive kinetic profiling of your antibody-antigen interactions. This SPR service can determine antibody binding affinities, association rates, and dissociation rates to support lead identification efforts and research and development pipelines. Inquire with our scientists about your antibody-antigen interaction project today.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics