Written by:
Written by:
Vanessa Yoon Calvelo, PhD
Published: December 22, 2022
The Great Debate of Antibody Discovery Platforms
The great debate on the use of in vivo versus in vitro sources and strategies for antibody discovery platforms continues to thrive among antibody research groups. On one side of the debate is the argument for non-animal-derived antibodies due to the technical advancements of current in vitro technologies, and the moral obligation to reduce animal usage. On the other side of the debate is the counterargument for animal-derived antibodies due to their better performance in affinity, specificity, and reduced immunogenicity risk.
Both sides have valid concerns, but the decision of which strategy to implement requires a detailed look at each type of antibody and the technologies used for their discovery and generation. In this article, we will compare in vivo and in vitro antibody discovery platforms with the hope to foster further discussions around these strategies for current and future workflows.
Traditional Antibody Discovery Technologies
The approach to antibody discovery and generation is a key factor to consider in the context of discovery campaigns. Traditional routes (Figure 1) that are still in use today include:
- Direct isolation from animal plasma
- Generation of hybridoma cells
- Screening of primary B cells
- Expression of recombinant antibody repertoires
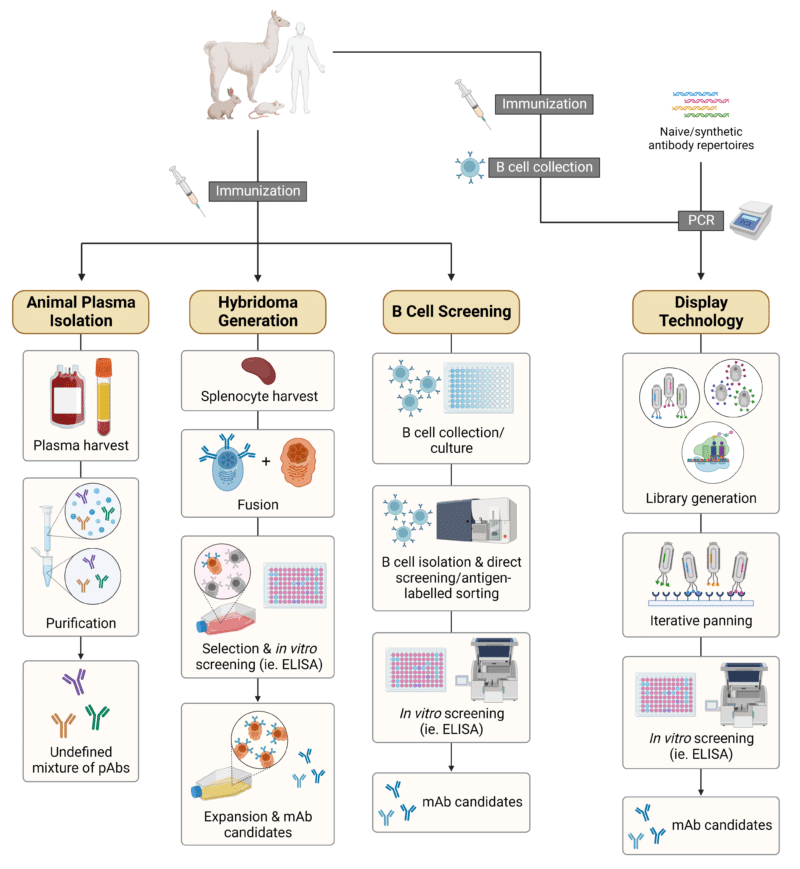
Figure 1. Schematic illustration of traditional strategies for antibody discovery and generation. Abbreviations: pAbs, polyclonal antibodies; mAb, monoclonal antibody.
Animal Plasma Isolation
Antibody isolation from animal plasma involves the animal’s immunization with an antigen (ie. full-length protein, peptide, stimulatory DNA) to drive the antibody response and maturation in vivo. By harvesting the animal plasma, removing non-antibody proteins, and antigen affinity purification, this approach yields the generation of an undefined mixture of polyclonal antibodies (pAbs). pAbs are widely known to have strong performance but suffer from batch-to-batch variability, loss of production animals, and difficulty with characterization. All other techniques in this article are centered on generating monoclonal antibodies (mAbs).
Hybridoma Generation
Hybridoma technology is an immortalization technique of short-lived antibody-secreting B cells. This process also begins by immunizing the animal with an antigen to provoke an immune response in vivo. The antibody-secreting B cells are then harvested typically from the animal spleen – though other organs and compartments have been harvested as well, including the bone marrow, lymph nodes, and peripheral blood mononuclear cells (PBMCs). Upon generation of hybridomas, by way of fusing antibody-secreting B cells with immortal myeloma cells, the hybridomas are cultured and its supernatant screened in vitro (ie. ELISA) for the presence of antigen-specific mAbs. Though hybridomas are accessible and widely used, they are often unstable and commonly express additional, productive heavy or light chains resulting in mAbs that are not monospecific.
B Cell Screening
Though hybridoma generation falls under the umbrella of ‘B cell screening’, there are alternative screening technologies that do not rely on the immortalization process. These alternatives are collectively referred to as ‘B cell screening’ and are distinct from hybridoma technology due to the direct application of primary B cells. B cells can be directly cultured, stained, and sorted by fluorescence-activated cell sorting (FACS), followed by in vitro screening for binding activity (ie. ELISA). Further downstream, the total collective of B cells can be sequenced using next-generation sequencing (NGS), known as B cell repertoire sequencing, or they can be sequenced individually, known as single B cell sequencing (described in the next section).
Like the hybridoma technology, B cell screening also relies on in vivo sources (ie. animal, human, transgenic animals) for mAb discovery and generation. The success of a discovery campaign depends on the sampling of B cells, the ability to isolate the critical antibody-secreting B cells, and the sustainability of the B cells ex vivo.
Check out our article: B Cells and Their Antibodies for a review on B cells and their differentiation.
Display Technologies
Improvements in current display technologies (phage, yeast, ribosome, bacterial, or mammalian display) have put a spotlight on this in vitro approach to antibody discovery and generation. Relying heavily on the linkage of genotype to phenotype, each given DNA sequence of an antibody fragment can be encoded within a phage or cell to be expressed on its surface. This underlying concept can be exploited to generate antibody repertoire libraries displaying millions of different antibodies, which can be screened iteratively, referred to as “panning”, against a given target antigen. Each sequential round of panning enriches the subsequent gene pool for the phages or cells that encode for the antigen binders.
Arguments could be made for display technologies to be entirely animal-free with the use of naïve antibody repertoires that do not reflect an immune system’s response to an antigen. Alternatively, display technologies can be used in combination with immunization, B cell sequencing, or hybridoma approaches to exploit DNA sequences of mAbs from an immunogenic response.
Proponents of display technologies argue that it is an effective way to iterate and test many mAb sequences to find the best binders. Detractors argue that since it is an in vitro approach, these synthetically derived sequences are likely to cause developability and immunogenicity challenges later on.
For an in-depth review on phage display, visit our article: Phage Display Antibody Discovery
Emerging Antibody Discovery Platforms and Technologies
To facilitate the ever-ongoing search for antibodies and antibody-derived formats, novel technologies have since emerged, providing further options in the approach to antibody discovery. A few emerging methodologies (Figure 2) include:
- De novo protein sequencing of polyclonal populations
- Sequencing and analysis of single B cell repertoires
- In silico discovery and generation with machine learning (ML)
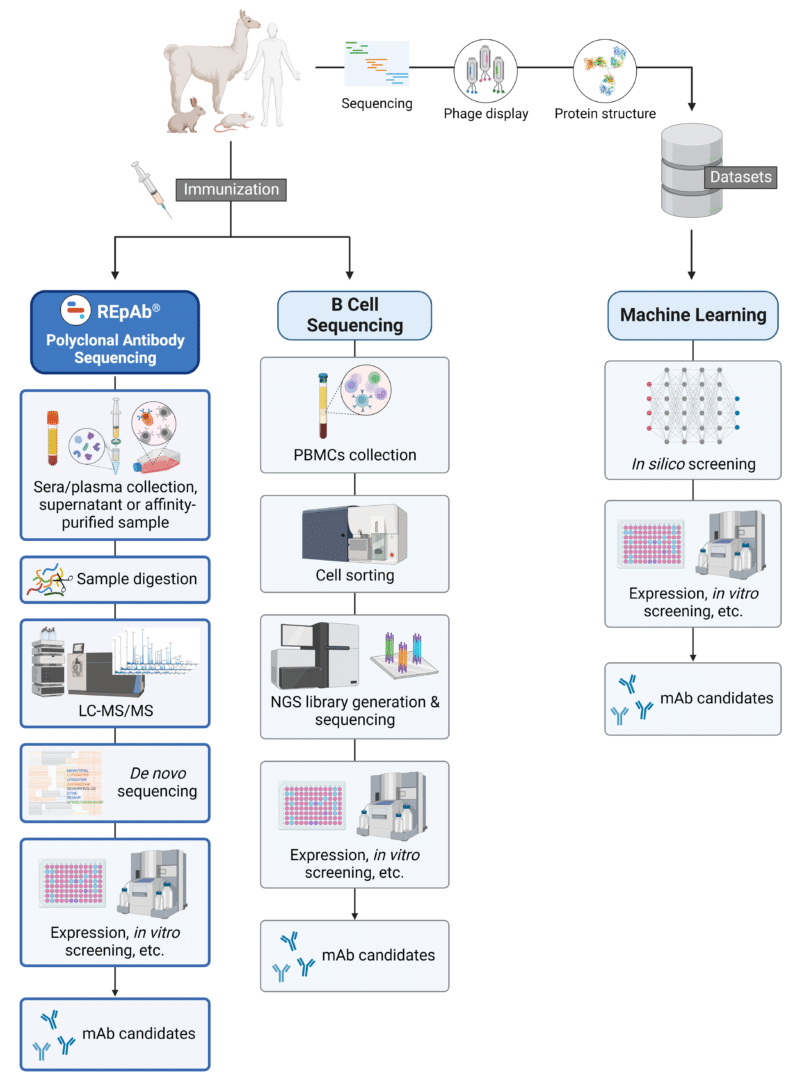
Figure 2. Schematic illustration of emerging strategies for antibody discovery and generation. Abbreviations: PBMCs, peripheral blood mononuclear cells; NGS, next-generation sequencing; mAb, monoclonal antibody.
Polyclonal Antibody Sequencing with REpAb®
Pioneered by Rapid Novor, polyclonal antibody sequencing with REpAb® is an emerging antibody discovery service that derives full-length mAb protein sequences directly from complex polyclonal populations. Antigen-specific pAbs are first collected from the serum of an immunized animal for affinity purification. The purified pAbs are digested into shorter peptides, which are analyzed via LC-MS/MS. Using proprietary ML-based bioinformatic algorithms, mAb protein sequences are then assembled de novo and identified for downstream applications (ie. recombinant expression, ELISA, HDX-MS epitope mapping, SPR kinetic analysis, etc).
REpAb® seamlessly integrates mass spectrometry-based proteomics and big data learning to identify and characterize the biologically relevant, circulating antibodies from their native source. This serum-only approach provides a path to in vivo antibody discovery and generation, avoiding the need to cull production animals or perform invasive medical procedures.
To learn more about how our REpAb® antibody discovery platform works, please refer to our article: What is Polyclonal Antibody Sequencing?.
Single B Cell Sequencing
An emerging technique among the suite of B cell screening technologies is single B cell sequencing. Primary B cells are collected and encapsulated with beads carrying barcoded oligos that anneal to single-cell total mRNAs upon lysis. These oligos are then amplified to produce a library for NGS. mAb clones are selected from the NGS data (mainly based on higher clonotype expansion) and subsequently synthesized, cloned, and expressed recombinantly.
Like other B cell screening technologies, single B cell sequencing also involves downstream in vitro screening, but relies on in vivo mAb discovery and generation.
For an in-depth review on B cell sequencing, visit our article: B Cell Sequencing and Polyclonal Sequencing Explained
ML-Assisted Discovery and Generation
ML has become a powerful tool in fostering scientific and technological advancements across many research fields, including antibody discovery. Current ML approaches for antibody discovery are primarily built on datasets derived from deep sequencing of antibody repertoires, screening of large phage display libraries, and/or scanning of 3D-structural predictions of antibody-antigen complexes.
Preliminary applications of ML for antibody discovery and generation have been demonstrated for in silico functional screening and developability optimization. However, the technology is still in its infancy and its success will depend on reliable training datasets obtained from both in vivo and in vitro sources.
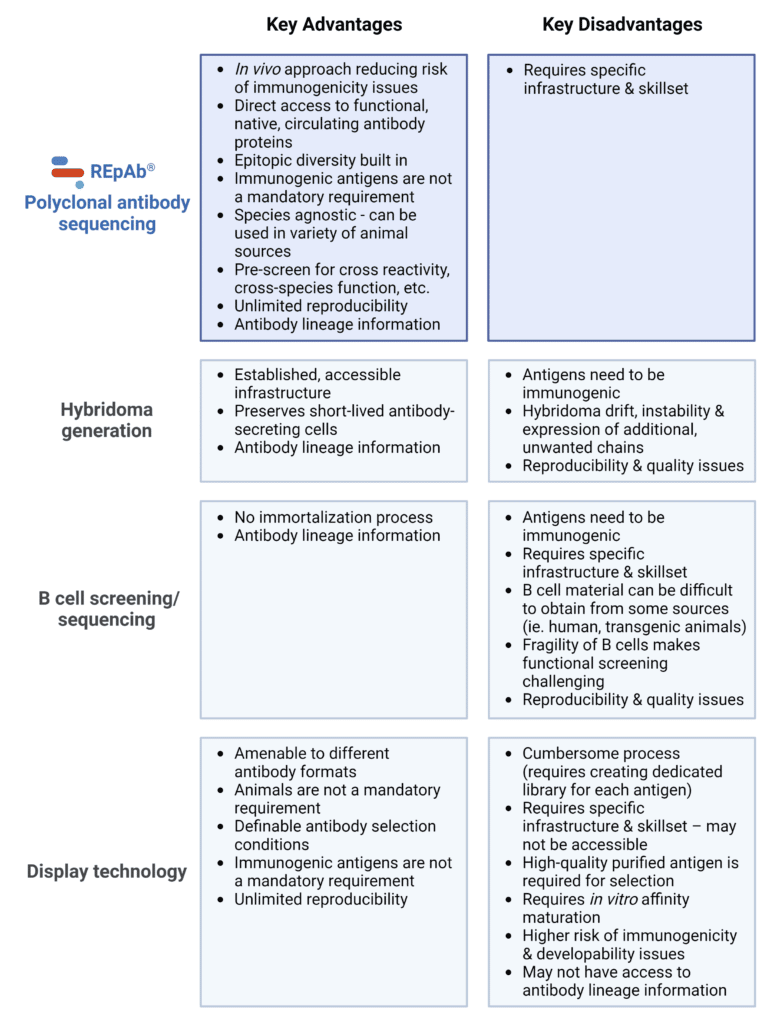
Antibody Discovery and Generation Technologies – Advantages and Disadvantages
It is abundantly clear that each strategy mentioned above can encompass both in vivo and in vitro experimental stages. Therefore, the approach to antibody discovery and generation should not be about advocating for one type of strategy and displacing the other. Rather, the focus should be on refining and applying these technologies towards generating antibody products with optimal properties and performance for their respective applications. A summary of the advantages and disadvantages of the four common methods for antibody discovery and generation is provided below (Table 1).
Table 1. Key advantages and disadvantages of REpAb® polyclonal antibody sequencing, hybridoma generation, B cell screening/sequencing, and display technology.
Antibody Discovery Services with Rapid Novor’s REpAb® Technology
One of the key benefits of the REpAb® technology is the ability to sequence native antibodies directly from a polyclonal protein sample. Our team has recently employed this strategy to successfully sequence the pAb population from an alpaca’s serum – a first-in-the-world achievement. Ten unique nanobodies were sequenced, recombinantly expressed, and revealed to exhibit high-affinity binding at the picomolar range.
Our REpAb® antibody discovery platform has been refined and advanced, allowing us to de novo sequence protein samples from other animals, including rabbit and goat. Our pAb sequencing technology supports the recombinant generation of biologically relevant antibodies, making batch-to-batch variability and irreproducibility a redundant afterthought. Talk to our scientists and learn more about how REpAb® antibody discovery service is the optimal strategy for your antibody discovery application.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics