
Written by:
Yuning Wang, PhD
Updated: January 26, 2023
(Published: March 15, 2022)
What are CDRs?
The acronym “CDR” stands for complementarity determining region, a variable sequence of amino acids that folds into loops capable of binding to an antigenic amino acid sequence, also known as an epitope (Figure 1).
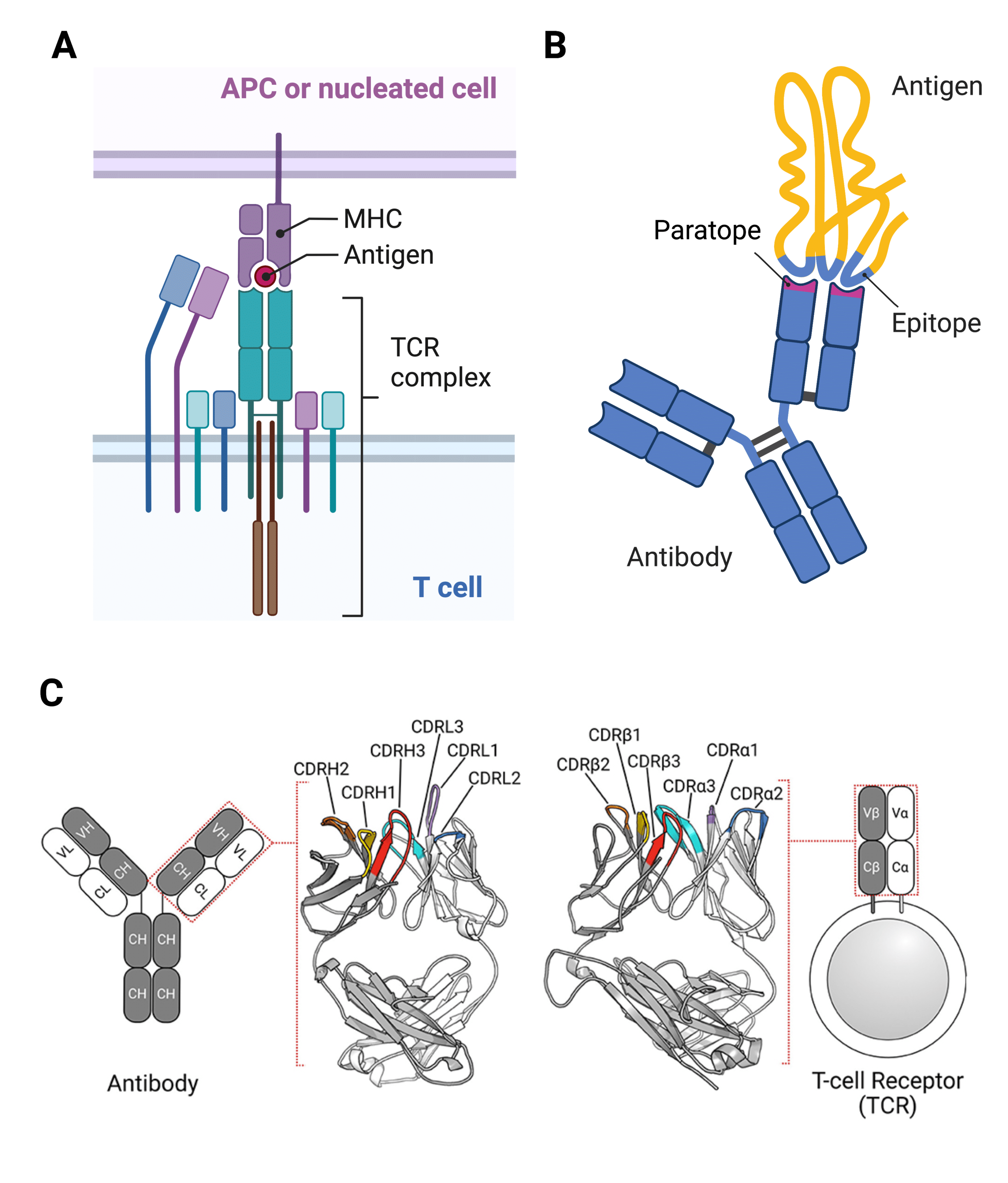
Figure 1. Complementarity Determining Regions (CDRs) are responsible for mediating the interactions between antigens presented by immune cells known as antigen-presenting cells (APCs) or other nucleated cells and the T cell receptor (TCR) part of the TCR complex of T cells (A), and between antigens and antibodies (B). The regions containing the CDRs are known as antigen-binding sites or paratopes (B). The antigenic area to which an antibody or TCR paratope binds is called the epitope (B). Both light and heavy chains of an antibody and the two extracellular chains of TCRs contain three CDRs each (C).
CDRs are found in both T cell receptors (Figure 1A) and antibodies (Figure 1B), where they are responsible for binding to antigens (Table 1).
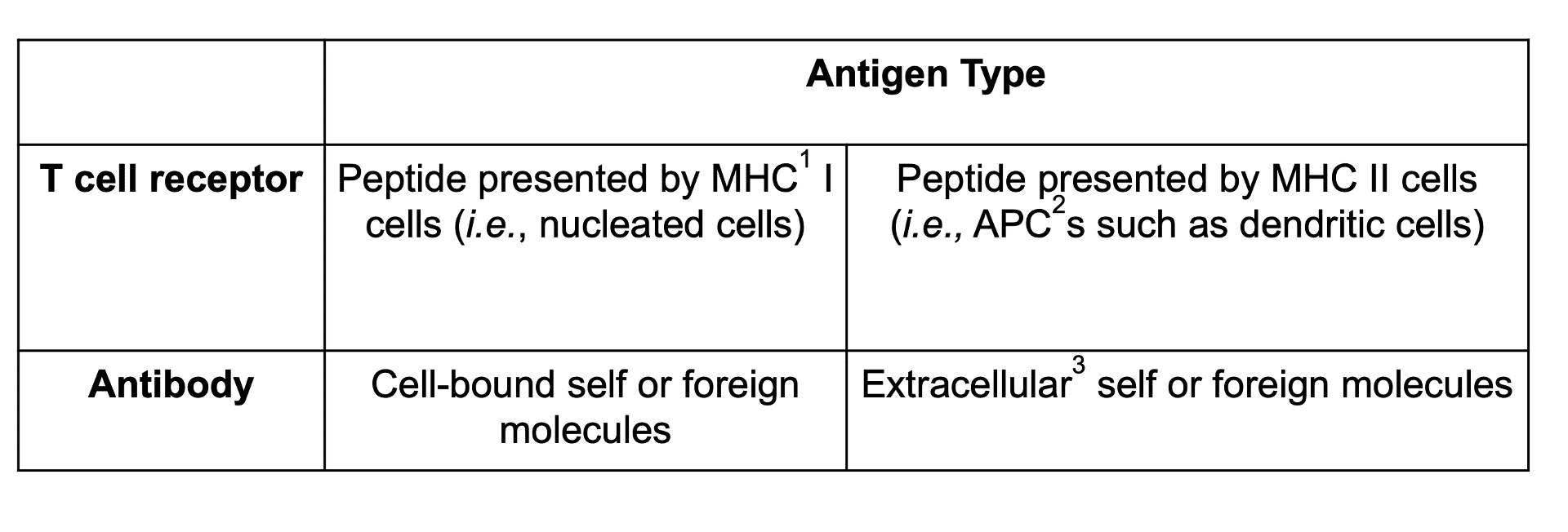
Table 1. Types of antigen recognized by T cell receptors and antibodies. 1MHC: Major histocompatibility complex. 2APC: Antigen-presenting cell. 3Though conventionally known to bind cell-bound or extracellular antigens, antibodies have been shown to also bind intracellular effectors outside of endosomal cycling to activate additional immune signaling pathways. More research is needed to understand this exciting development.
Antibody CDRs
When we think about antibodies, particularly for monoclonal antibodies (mAbs) used as therapeutics or diagnostics, we often picture the classic Y-shaped protein immunoglobulin G (IgG). To illustrate CDRs in antibodies, this article will be focusing on the structure of a typical IgG. Please consult our other article for other types of antibodies.
How Many CDR Loops are in the IgG Antibody Structure?
IgG antibodies are composed of two identical peptide chain pairs: heavy chain and light chain. Thus, IgG antibodies are heterotetrameric proteins. Both the light and heavy chains are made of variable (V) and constant (C) regions; each variable region contains three CDR loops that form the majority of the antigen-binding site. Outside of the CDR loops, framework sequences stabilize the loop structure (Figure 2).
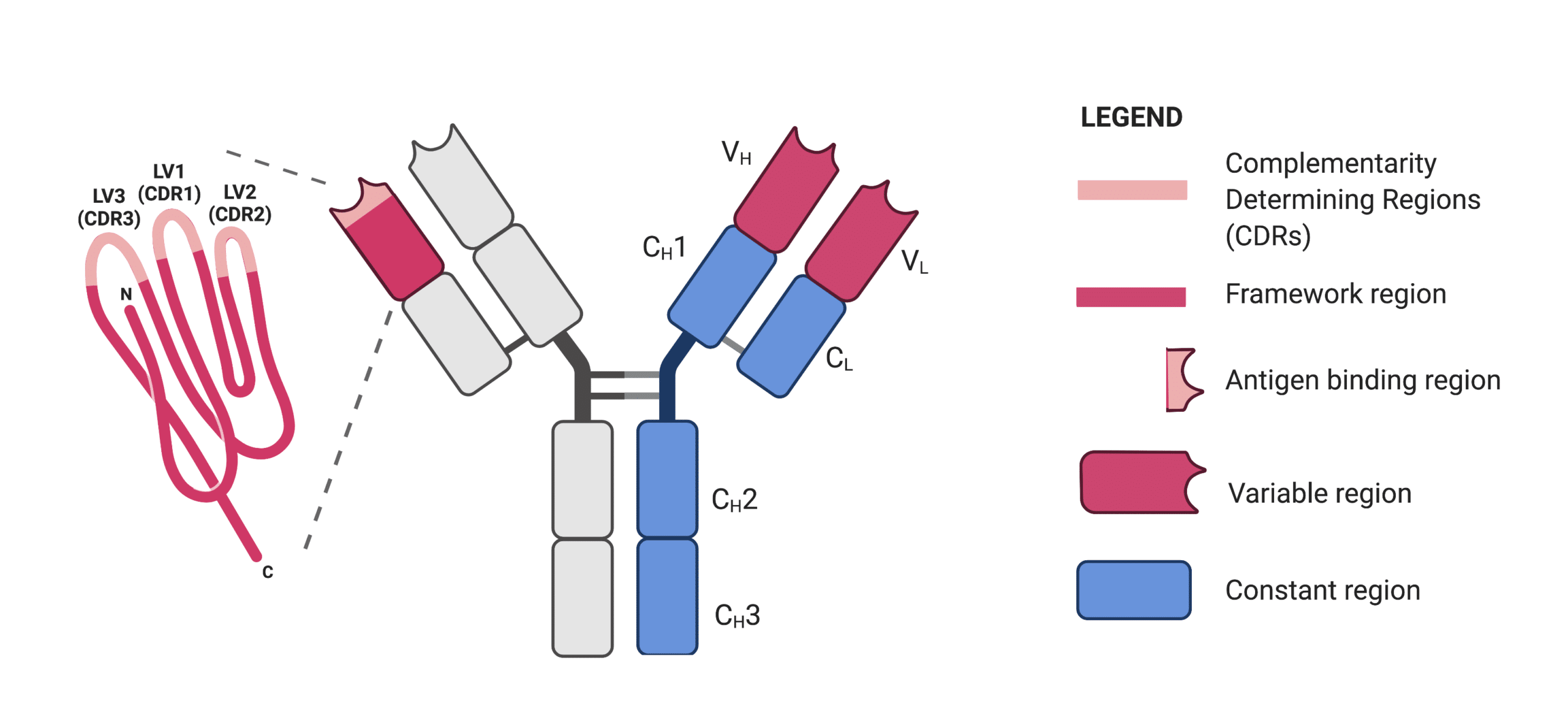
Figure 2. Schematic representation of an IgG antibody. The enlargement shows the variable domain of the light chain (LV) consisting of three CDR loops and framework region.
Variability in Antibody CDR Sequences
Because antibodies’ CDRs are highly variable, it can prove difficult to identify their exact amino acid sequence. Essentially, CDRs are novel protein sequences. Routine identification of proteins is often executed via genomics and proteomics methodologies that require database searches. Since antibody sequences are rarely found in databases, more advanced techniques are required.
This article will review the advantages of using de novo protein sequencing to identify CDRs by sequence, as well as depict, in a specific research case study, how researchers used this method to successfully accomplish the accurate identification of CDRs. Furthermore, this article will provide further resources to help those studying proteins to enhance their workflows with next generation protein sequencing.
De Novo Protein Sequencing as a Tool for Identifying CDRs Sequences
De novo protein sequencing is a well-established procedure that relies on mass spectrometry to sequence proteins – the technique is well described in our previous report and has been advanced significantly in the last 5 years. This technique is advantageous as it does not require information on the amino acid sequence of peptides to be already known, circumventing limitations of methods that rely on protein sequence databases.
De novo protein sequencing produces results directly from mass spectrometry data which makes it especially useful for identifying highly variable CDR’s sequences. This type of sequencing can accurately identify each amino acid in each position. That being said, the technique naturally generates the entire antibody sequence. Therefore, overlapping peptide amino acid sequences from the CDRs provide multiple confirmations of each amino acid position in the CDR, and also orient the CDR sequence within the complete antibody sequence.
For more information on different sequencing strategies, including de novo protein sequencing, please see the article Antibody Protein Sequence Analysis Using Mass Spectrometry.
Applying De Novo Protein Sequencing to Identify CDRs Sequences
Recently, the Zubarev lab at the Karolinska Institutet, Sweden, successfully exploited Rapid Novor’s REmAb® de novo sequencing technology to develop a novel, high sensitivity, quality and throughput immunoassay for Alzheimer’s disease. Monoclonal antibodies were first generated in hybridoma cells, but the Zubarev lab needed to use de novo sequencing to fully understand the amino acid sequence of the CDR regions – as it turns out, a single amino acid position was vital to binding.
This is one of many case studies that have benefitted from fully characterizing the CDR sequence using antibody sequencing. Rapid Novor alone has successfully completed over 5000 antibody sequencing projects including confident assignment of CDRs.
Annotation Schemes for Identifying CDRs by Sequence
Having obtained the full-length sequence of an antibody, it is important to identify which portions of the sequence correspond to the CDR loops and which to the framework regions. Doing so implies prediction of secondary structure – fortunately antibodies have a fairly predictable secondary structure so several standard schemes have emerged for annotating the CDR regions of a linear protein sequence, the most common of which is referred to as Chothia. Dondelinger et al. have presented a comprehensive review of the commonly used annotation schemes and the methodology behind them:
- Chothia – a structure-based numbering scheme used by Rapid Novor in annotation of REmAb antibody sequencing reports
- Kabat – the most widely adopted sequence-based scheme for numbering antibody residues
- Martin – the most recent version of the Chothia numbering with correction on insertion sites in framework regions
- Glefand – a numbering method that provides comparison of secondary structures between aligned sequences
- IMGT – a standardized numbering system based on alignments of sequences from a complete reference gene database (Figure 3)
- Honnegers – the “Aho” numbering scheme based on known three-dimensional structures of immunoglobulin domain
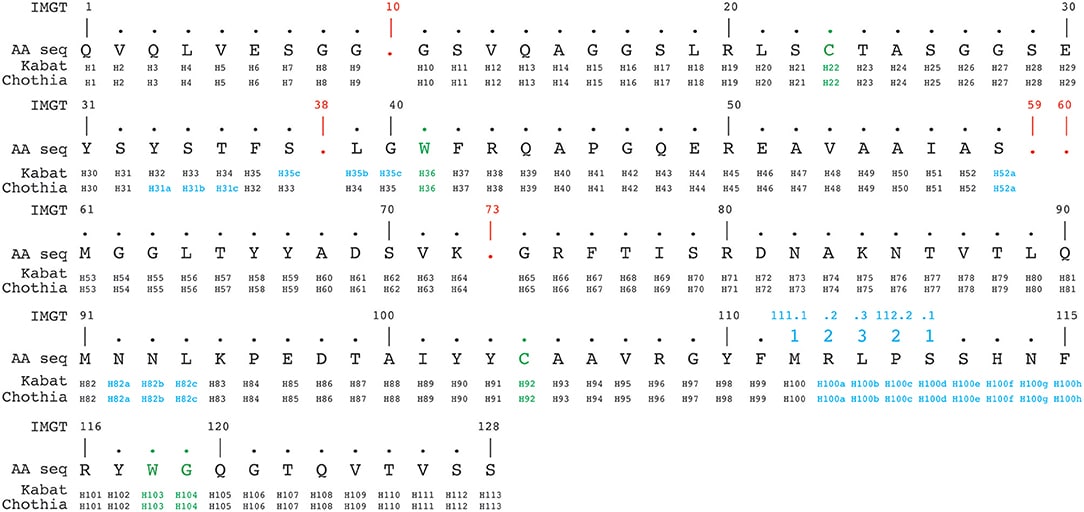
Figure 3. An example of Alignment of a nanobody sequence according to Kabat, Chothia, and IMGT numbering schemes. Green: conserved cysteine-cysteine-tryptophan triads forming disulfide bonds. Red: absent residues (‘gaps’) according to the the IMGT numbering scheme. Blue: insertion positions. Image credit: https://www.frontiersin.org/articles/10.3389/fimmu.2018.02278/full#h13
These schemes mostly agree, but you can expect different/additional amino acid positions are considered important by one scheme or the other. It is therefore important to not underestimate the importance of amino acids in ‘framework’ regions.
Additionally, here are several free online software for annotating CDR based on sequence:
Antibody Sequencing Service
An expert in the field of protein sequencing, Rapid Novor can provide de novo protein sequencing services to allow high quality identification of CDRs by sequence. Rapid Novor’s recognized scientists and technicians work to pioneer solutions in protein sequencing, prioritizing client needs. Research involving identifying CDRs by sequence can benefit from de novo protein sequencing and the solutions provided by Rapid Novor to accomplish this. If you would like to learn more about identifying CDR loops by sequence or have any questions about applications for your unique protein sequencing purposes, please do not hesitate to contact a member of our team today.
Resources:
https://www.rapidnovor.com/antibody-protein-sequence-analysis-using-ms/
https://pubs.acs.org/doi/10.1021/acs.jproteome.1c00169
https://www.frontiersin.org/articles/10.3389/fimmu.2018.02278/full
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2396520/
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

