Written by María Gerpe, PhD
Written by María Gerpe, PhD
July 7, 2021
(Updated: May 3, 2023)
Isoleucine
Isoleucine is a non-polar, non-charged at pH 7.0, non-aromatic, branched-chain amino acid that cannot be synthesized by humans. As a result, it is one of the nine essential amino acids along with histidine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine.
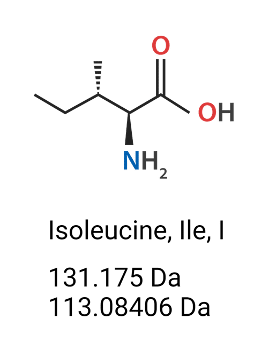
Figure 1. Isoleucine (Ile, I) amino acid’s chemical structure, molecular weight (Da) (mass which includes the extra H2O) and monoisotopic residue mass (Da) (does not include the hydroxyl group of the carboxylic acid nor one of the hydrogens on the amine).
Like all amino acids, it is composed of an alpha-amino group, and alpha-carboxylic group. Because of this structure, as amino acids link with each other to form a protein, all proteins are defined by an N-terminal end, and a C-terminal end.
Isoleucine is encoded by AUU, AUC, and AUA codons. Isoleucine is frequently present in the antigen-binding regions of antibodies.
Leucine
Similarly to isoleucine, leucine is another non-polar, non-charged at pH 7.0, non-aromatic, branched-chain, essential amino acid. It is encoded by UUA, UUG, CUU, CUC, CUA, and CUG, and it is structurally similar to isoleucine. For this reason, both isoleucine and leucine are difficult to distinguish via standard mass spectrometry means.
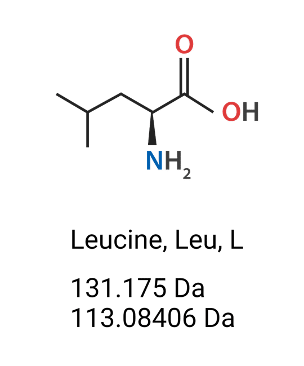
Figure 2. Leucine (Leu, L) amino acids’ chemical structure, molecular weight (Da) (mass which includes the extra H2O) and monoisotopic residue mass (Da) (does not include the hydroxyl group of the carboxylic acid nor one of the hydrogens on the amine).
Isoleucine and Leucine Have the Same Mass
Because they share the same mass, isoleucine and leucine are known as isobaric amino acids. Conventional mass spectrometry-based proteomics cannot be easily used to distinguish between isoleucine and leucine. However, in 2016, Rapid Novor became the first to commercially introduce a service, w-ion isoleucine-leucine determination (WILD® ), to distinguish between Ile and Leu in de novo antibody sequencing.
Why is it important to distinguish between Isoleucine and Leucine in Antibodies?
Each antibody has six complementarity-determining regions (CDRs): three on each heavy and light chain, perhaps the most important sites for antibody/antigen interaction. Incorrect identification of a single amino acid in the CDRs can lead to expressing an antibody with unexpected binding and biological activity. This is particularly the case for CDR-H3, which is the most variable and unique of all CDRs (1). For this reason, it is crucial to be able to identify the sequence of the CDRs with 100% certainty, including differentiating between Ile and Leu residues. The latter is especially important because Ile and Leu are frequently found in these hypervariable regions (2).
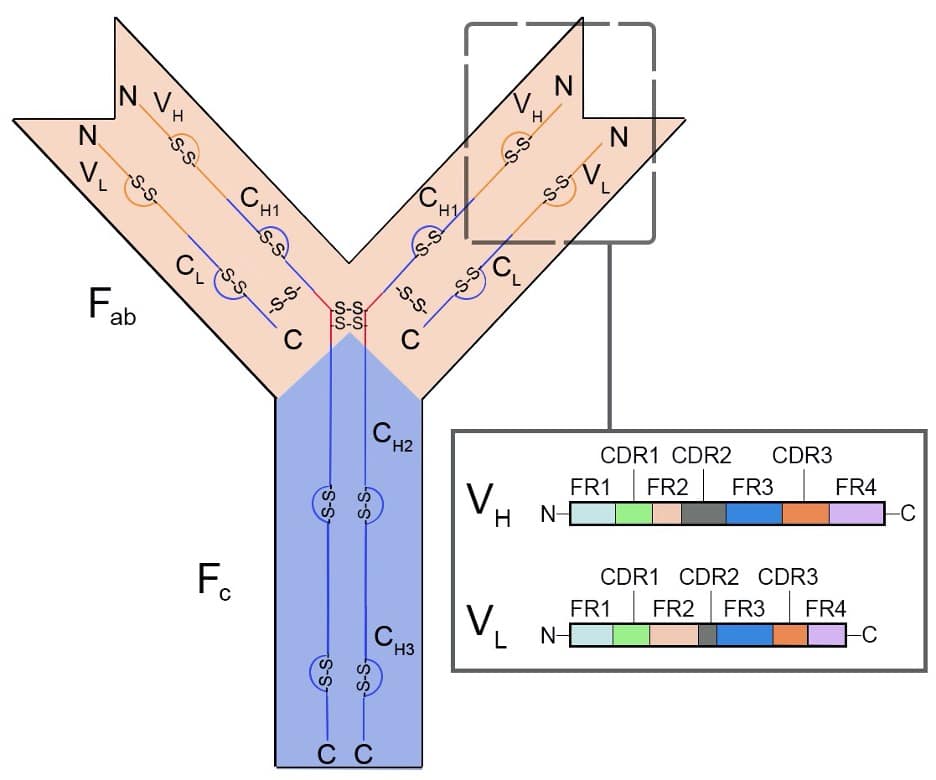
Figure 3. Representative diagram of protein structure of an IgG class antibody. From N-terminus to C-terminus, the antigen-binding region (Fab) contains a variable (orange) and constant (blue) region for both heavy (VH and CH1-3) and light chains (VL and CL), respectively. The variable regions (VH and VL) (inset box) contain the complementarity determining regions (CDRs1-3) interspersed through framework regions (FR1-3). The Fab region is separated from the crystallizable fragment (Fc) domain by a disulphide bridge or hinge (red) with IdeS digestion. The Fc is composed of constant domains from the two heavy chains.
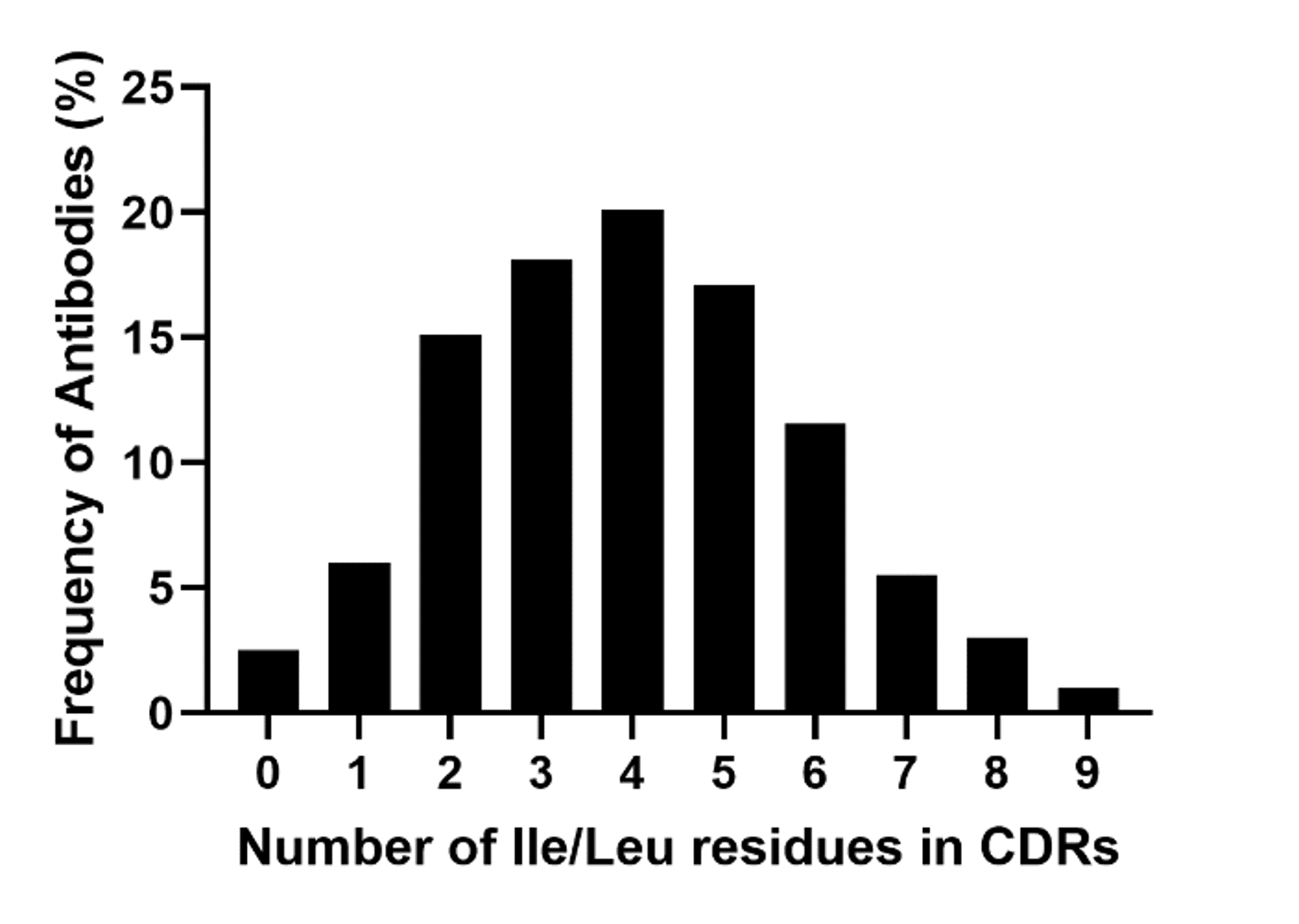
We have found that isoleucine and leucine are frequently observed in monoclonal antibodies. In 2017, we sequenced 200 monoclonal antibodies, and found that CDRs with no isoleucine or leucine residues are very rare (3). Ninety-seven percent of antibodies will contain at least one isoleucine or leucine residue in their CDRs, and more than half of the antibodies we sequenced using our popular REmAb® sequencing service contained at least four isoleucine or leucine residues in their CDRs. Moreover, approximately 10% of sequenced antibodies contained more than seven isoleucine or leucine residues.
Figure 4. Frequency of antibodies expressing isoleucine or leucine in their complementarity determining regions (CDRs). A total of 200 monoclonal antibodies were sequenced using our de novo protein sequencing platform (REmAb®).
Take a moment to imagine. If an antibody had a CDR containing nine isoleucine/leucine positions, you would be required to express 512 forms just to confirm the full antibody sequence. Why resort to guesswork when you can have certainty with WILD®?
Other isobaric amino acids like isoleucine and leucine
In addition to WILD®, REmAb® can also identify other isobaric residues. By taking into account proteolysis and peptide fragmentation patterns, our machine learning software assigns confidence scores to each amino acid and peptide derived from mass spectral data. We also digest our samples with multiple proteases to yield many overlapping peptides (4) that can help us verify the entire protein sequence with great accuracy and redundancy.
How does w-ion isoleucine and leucine determination (WILD®) work?
To elucidate the mass of proteins, digested peptides are run through a mass spectrometer where they undergo collisions that induce fragmentation of the peptide backbone, generating N- and C-terminal ions. C-terminal z-ions contain the side chains of the peptide backbone. When fragmentation of the z-ion sidechains occurs, w-ions are generated. Though isoleucine and leucine are isobaric, their w-ions are not.
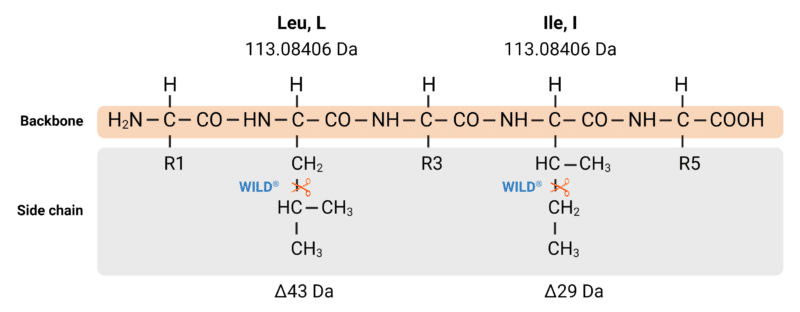
Figure 5. Although isoleucine and leucine share the same mass (113.08406 Da), our WILD® technology employs controlled fragmentation in a mass spectrometer to cleave off different parts of their side chains. This controlled fragmentation yields different w-ions, which are used to distinguish I and L.
The w-ion was first reported in 1987 by Johnson, Martin, and Biemann (5,6). However, it was mostly known in academic circles. Rapid Novor became the first to commercially offer a w-ion-based isoleucine from leucine determination in antibody sequencing. At Rapid Novor, we employ proprietary protein chemistry and state-of-the-art Orbitrap Fusion instruments that can perform electron-transfer high-energy collision dissociation (EThcD) (7). EThcD of z-ions yields w-ions. Our protein chemistry allows us to consistently and successfully determine all I/Ls in our clients’ samples. With WILD®, we uncover I and L residues accurately and with high throughput.
Having been the first ones to introduce WILD® for antibody sequencing commercially, we have successfully completed the most isoleucine/leucine determination projects with unparalleled accuracy and throughput. With WILD®, you can keep working with confidence.
To learn more about our antibody sequencing services, contact us.
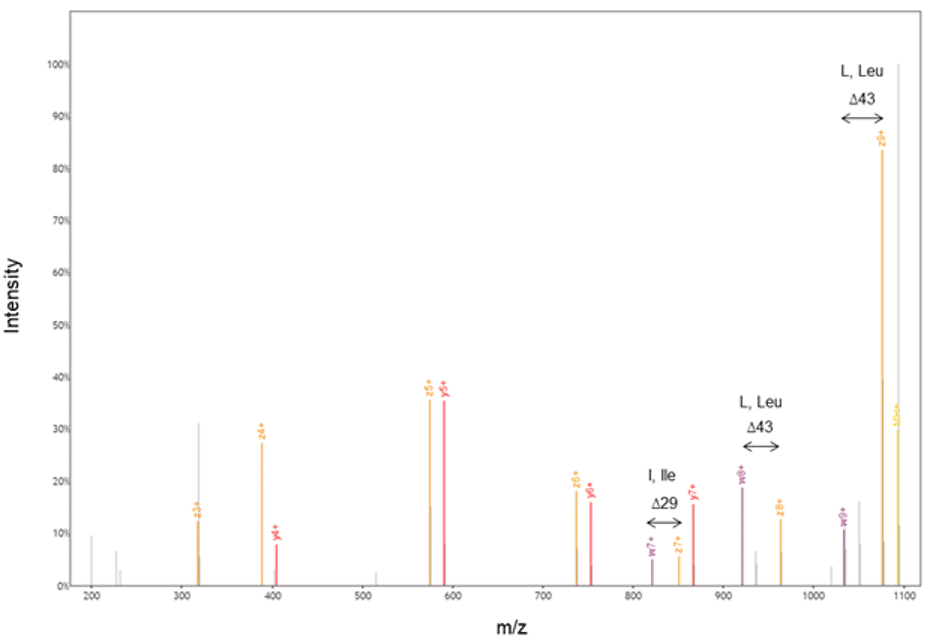
Figure 6. Our proprietary protein chemistry and EThcD allowed us to generate w- and z-ions; our accurate and automated machine learning algorithms then calculated the difference between these ions to annotate the sequence.
References
- Sela-Culang, I., et al., The Structural Basis of Antibody-Antigen Recognition. Frontiers in Immunology, 2013. 4.
- Gonzalez-Munoz, A., et al., Tailored amino acid diversity for the evolution of antibody affinity. MAbs, 2012. 4(6): p. 664-72.
- McDonald, Z.L., Q; Stajduhar, A; Taylor, P; Krieger, JR; Ma, B, Large Scale Study of the W-ion Isoleucine and Leucine Determination (WILD™) Method in Antibody De Novo Protein Sequencing, in 66th American Society for Mass Spectrometry. 2018: San Diego.
- Le Bihan, T., et al. Increased De Novo Protein Sequencing Coverage with Optimal Protease Cocktail. in American Association for Mass Spectrometry. 2019. Atlanta.
- Ma, B., Novor: real-time peptide de novo sequencing software. Journal of the American Society for Mass Spectrometry 2015. 26(11): p. 1885-1894.
- R. S. Johnson, S. A. Martin and K. Biemann, “Novel Fragmentation Process of Peptides by Collision-Induced Decomposition in a Tandem Mass Spectrometer: Differentiation of Leucine and Isoleucine,” Analytical Chemistry, vol. 59, no. 21, pp. 2621-2625, 1987.
- S. S. Zhokhov, S. V. Kovalyov, T. Y. Samgina and A. T. Lebedev, “An EThcD-Based Method for Discrimination of Leucine and Isoleucine Residues in Tryptic Peptides,” Journal of the American Society for Mass Spectrometry, vol. 28, no. 8, pp. 1600-1611, 2017.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics