 Written by Yuning Wang, PhD
Written by Yuning Wang, PhD
August 1, 2021
What is DNA Sequencing?
DNA sequencing is the process of determining the precise order of four nucleotides bases—adenine (A), guanine (G), cytosine (C), and thymine (T)—that make up the DNA molecule. From Sanger sequencing to next-generation sequencing (NGS), DNA sequencing’s accessibility and ease of use make it one of the most widely used technologies in life sciences.
After sequencing the first protein – insulin, Frederick Sanger and colleagues developed a DNA sequencing method named Sanger sequencing after its lead inventor in the 1970s. Sanger Sequencing is the earliest and most established DNA sequencing method. It is a chain termination method by selectively incorporating radioactively or fluorescently labeled dideoxynucleotides during in vitro DNA replication. The recent set of DNA sequencing technologies that emerged since the early 2000s are collectively referred to as next generation sequencing (NGS) (Figure 1). The fundamental concepts behind NGS technology are similar to Sanger sequencing. What makes NGS different mainly includes increased data output, improved efficiency, and the ability to sequence whole genomes and low-frequency variants. These improvements were made by the integration of automated sequencing strategies, advanced data acquisition methods, and bioinformatics tools. NGS allows for massively parallel sequencing of millions to billions of DNA nucleotides, and greatly reduces cost and time of obtaining large-scale genomic data.
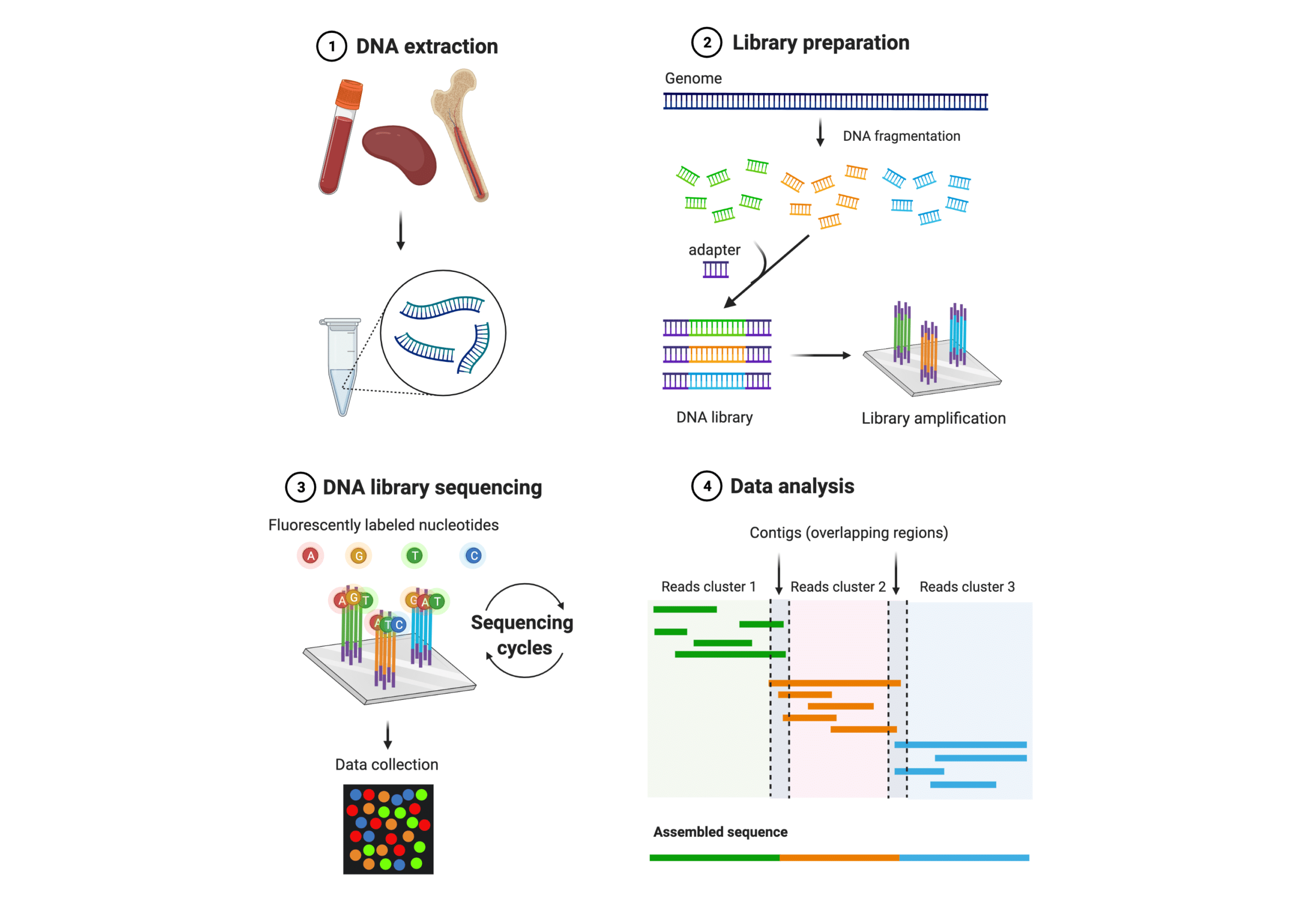
Figure 1. General workflow of NGS
DNA Sequencing in Antibody Development
DNA sequencing technologies, especially NGS, are extensively applied in the process of antibody discovery and development. Techniques like hybridoma sequencing and phage and yeast display technology that rely on NGS or nucleotide sequencing methods have strengthened the development of antibody therapeutics and diagnostics.
Hybridoma sequencing, a type of nucleotide sequencing, can obtain sequence information of monoclonal antibodies produced by hybridoma cell lines, widely used for the production of humanized antibodies, and in the initial screening and lead selection. Display technologies rely on NGS to generate libraries of antibody sequences that are then expressed or displayed in large numbers on the surface of phages, yeast, or bacteria, allowing the rapid identification of antibodies and fragments that bind target molecules.
NGS may also be used for recombinant expression of antibody clones through two means: single B cell sequencing or B cell repertoire sequencing (BCR), acting as a powerful tool to analyze large-scale sequence data of antibody repertoires in response to particular antigens or diseases directly from blood B cells.
Single B cell sequencing relies on the individual culturing of B cells prior to sequencing, retaining information on light and heavy chain pairing. BCR sequencing depends on the bulk mRNA extraction of B cells, particularly from the spleen. Both help researchers gain important knowledge of the humoral adaptive immune response to identify therapeutics and monitor disease in clinical settings.
Limitations of DNA Sequencing
Certain limitations exist within the above mentioned DNA sequencing methods. Hybridoma sequencing can often be failed due to the fragility and instability of hybridoma cell lines; Phage/yeast display requires high cost and laborious process, and may generate antibodies with incorrect pairing of heavy-light chains. Besides, antibodies selected may not show consistent affinities in vivo that are achieved in vitro during affinity maturation, causing delays in clinical development.
In regards to B cell sequencing, as it is restricted to circulating B cells, it only represents a minority of the total B-cell population. Only a small part of the BCR on the surface of B cells will be secreted into the serum to form soluble functional antibodies. Therefore, B cell sequencing cannot accurately capture nor represent the circulating antibodies at serological levels. Finally, nucleotide sequencing methods are incapable of examining post-translational modifications (PTMs), which are greatly important for binding, stability, and half-life.
What is De Novo Protein Sequencing?
Compared with DNA, proteins are sometimes more suitable to be used as biomarkers to track and detect diseases, and as therapeutic molecules for medical research and development, so the need to sequence proteins is of great importance. De novo protein sequencing is the method in which the amino acid sequence of a protein is directly determined without prior knowledge of its DNA sequence. The technology can be traced back to half a century ago when Edman sequencing was developed to sequence short peptides. Over the past few decades, with advancements in mass spectrometry-based proteomics, Edman sequencing has been largely taken over by the new generation of de novo sequencing built on tandem mass spectrometry.
The central idea of protein de novo sequencing by mass spectrometry is to determine the mass of an amino acid residue in a protein by measuring the mass difference between two fragment ions from tandem mass spectra. In such a way, the entire protein sequence can be inferred by assigning each residue along the protein backbone (Figure 2). It is particularly useful when the cDNA or the original cell line of a protein is not available, and could be done without use of databases compared to traditional protein sequencing.
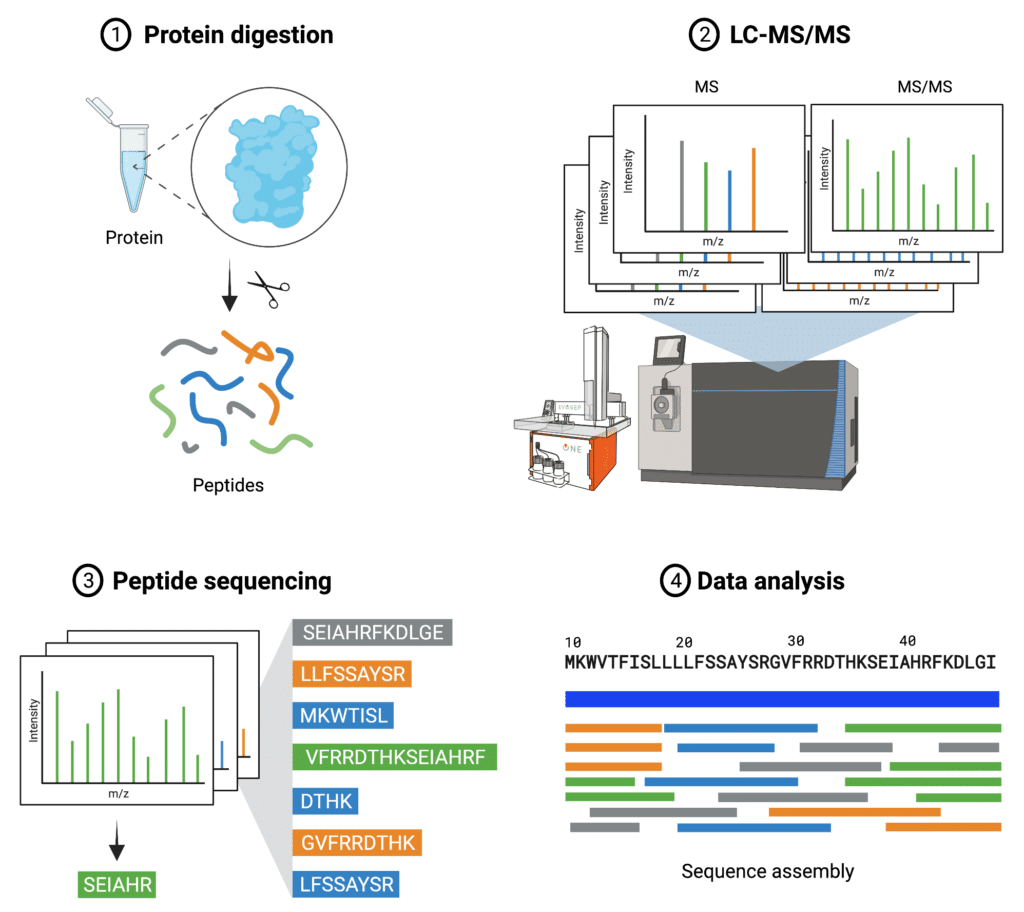
Figure 2. Steps of de novo protein sequencing.
De Novo Protein Sequencing of Monoclonal and Polyclonal Antibodies at Rapid Novor
Our REmAb® antibody sequencing service at Rapid Novor provides an approach for sequencing monoclonal antibodies with mass spectrometry. We have developed an advanced and proprietary algorithm for protein de novo sequencing with an emphasis on sequencing the CDR regions of antibodies correctly. REmAb® directly analyzes the antibody protein (only 0.1 mg required) without the need for cell lines or DNA information with 100% coverage and accuracy in s short turnaround time. To date, we have successfully sequenced more than 3000 monoclonal antibodies, including antibody reagents, therapeutics, and newly discovered antibodies. The sequences have been used in a wide range of applications for antibody development from antibody engineering, therapeutic developability assessment, to lead selection.
Currently available de novo protein sequencing techniques are still exclusive for sequencing single proteins like monoclonal antibodies, rather than protein mixtures containing a variety of heterogeneous proteins. This limit has recently been broken by Rapid Novor. For the first time in the world, using REpAb® – antibody discovery service, our team successfully de novo sequenced antibodies from a polyclonal mixture without the use of DNA or other nucleotide sequencing technology. The antibody discovery platform directly analyzes a polyclonal sample, or the plasma or serum of an immunized animal or convalescent plasma of a patient. By using our de novo protein sequencing software, full-length sequences of dominant monoclonal antibodies can be derived from the mixture. In such a way, REpAb® reflects more accurate and specific immune repertoires compared to BCR sequencing as it’s based on relative abundance information, and reports on the end-point product (i.e., IgGs).
Advantages of Combining De Novo Protein Sequencing & DNA Sequencing
De novo protein sequencing can offer effective complements to DNA sequencing in many applications, especially in antibody repertoire analysis. By combining the proteomics approach like polyclonal antibody sequencing with B cell sequencing, the advantages of the two methods can be fully employed. This is what our REpAb antibody discovery platform aims at. Upon animal immunization, de novo protein sequencing and B cell sequencing can be performed in parallel. The proteomic data can be interpreted in reference to transcriptomics databases derived from B cell sequencing, which helps narrow down the large amounts of sequences to the most functional ones (Figure 3). This not only allows for a more comprehensive and accurate analysis of the immune response, but also greatly expedites the process of antibody-based therapeutic discovery and development.
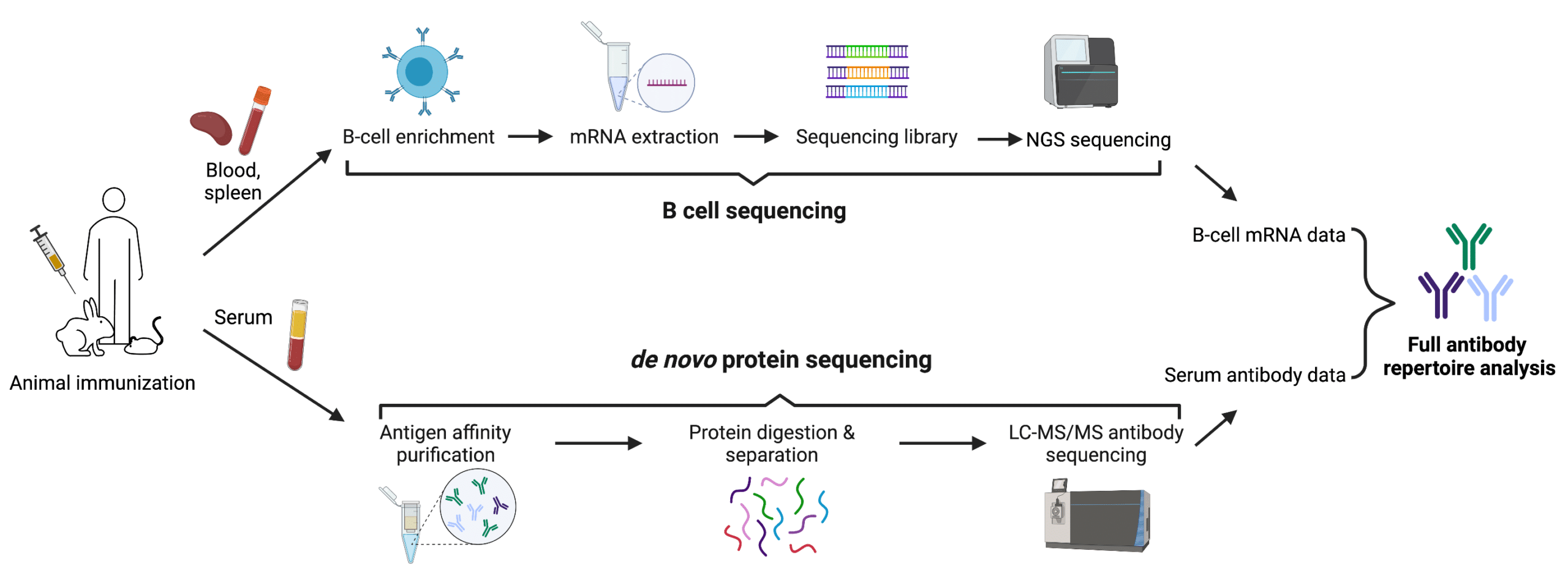
Figure 3. Combination of de novo protein sequencing and DNA sequencing in antibody repertoire analysis.
Additionally, there are other circumstances that de novo protein sequencing could be considered over DNA sequencing, which include the following:
- To examine PTMs or glycosylation of antibodies
- When the cell line of the antibody is lost or inaccessible
- For non-model species lack of PCR primers for DNA sequencing
- To design a series of engineered antibodies by CDR grafting
- To monitor the real-time status of antibodies or proteins in patients
- To develop biosimilars from off-patent antibodies
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

