Zac McDonald1, Qixin Liu1, Mingjie Xie1, Paul Taylor1, and Bin Ma1,2
1Rapid Novor, Inc, Kitchener, Ontario, Canada
2University of Waterloo, Waterloo, Ontario, Canada
Abstract
Monoclonal antibodies (mAbs) are widely used in research, diagnosis, and pharmaceutical purposes. Lately, the relatively lower quality of research-purpose mAbs is a point of concern within the research community. The problems include the lack of validation and low reproducibility. As most of the validation is done by immunoassay, the extent of protein contamination, including undesired antibody forms, remains unknown.
Mass spectrometry-based protein sequencing technology enables the discovery and characterization of undesired antibody forms in mAb reagents. In this study, we examine the existence of a second light chain in addition to the expected primary light chain. To our surprise, a significant portion of the mAbs we studied included a second light chain that can be completely or partially de novo sequenced via mass spectrometry.
Key Takeaways
- De novo protein sequencing is capable of identifying additional antibody chains
- De novo protein sequencing can decode antibody sequences regardless of the isotype or the host species
- Secondary light chains may affect antibody binding and contribute to poor reproducible results
- Reproducibility is a cornerstone of the immunoreagent industry
- De novo protein sequencing can be used to secure reproducibility of immunoreagents
Introduction
The quality of monoclonal antibodies in research has been a recent point of contention among researchers1,2. Some of the problems highlighted has been the presence of contamination and additional antibody chains.
De novo protein sequencing technology can be employed to elucidate undesired antibody forms in immunoreagents for commercial or in-house use.
This study explored the use of mass spectrometry and Big Data-based bioinformatics in de novo sequencing of 80 commercial, intended-for-research antibodies.
Materials & Methods
Each antibody sample was digested with six different enzymes, Trypsin, Lys C, Chymotrypsin, Pepsin, Asp N, and Elastase (Promega, WI, US). Tandem mass spectrometry (MS/MS) data were collected with an Orbitrap Q Exactive (Thermo Fisher, CA, US) for each digest.
Novor software3 was used to de novo sequence each peptide. The protein sequencing was assembled with the REmAb® sequencing platform (Rapid Novor, Kitchener, ON, Canada) (Figure 1).
After sequencing of the first pair of heavy and light chains for each antibody, the software then sequenced a second light chain from the data.
A second light chain was reported as found if it had significant sequence coverage based on liquid chromatography (LC)-MS/MS data and was sufficiently different from the first one.
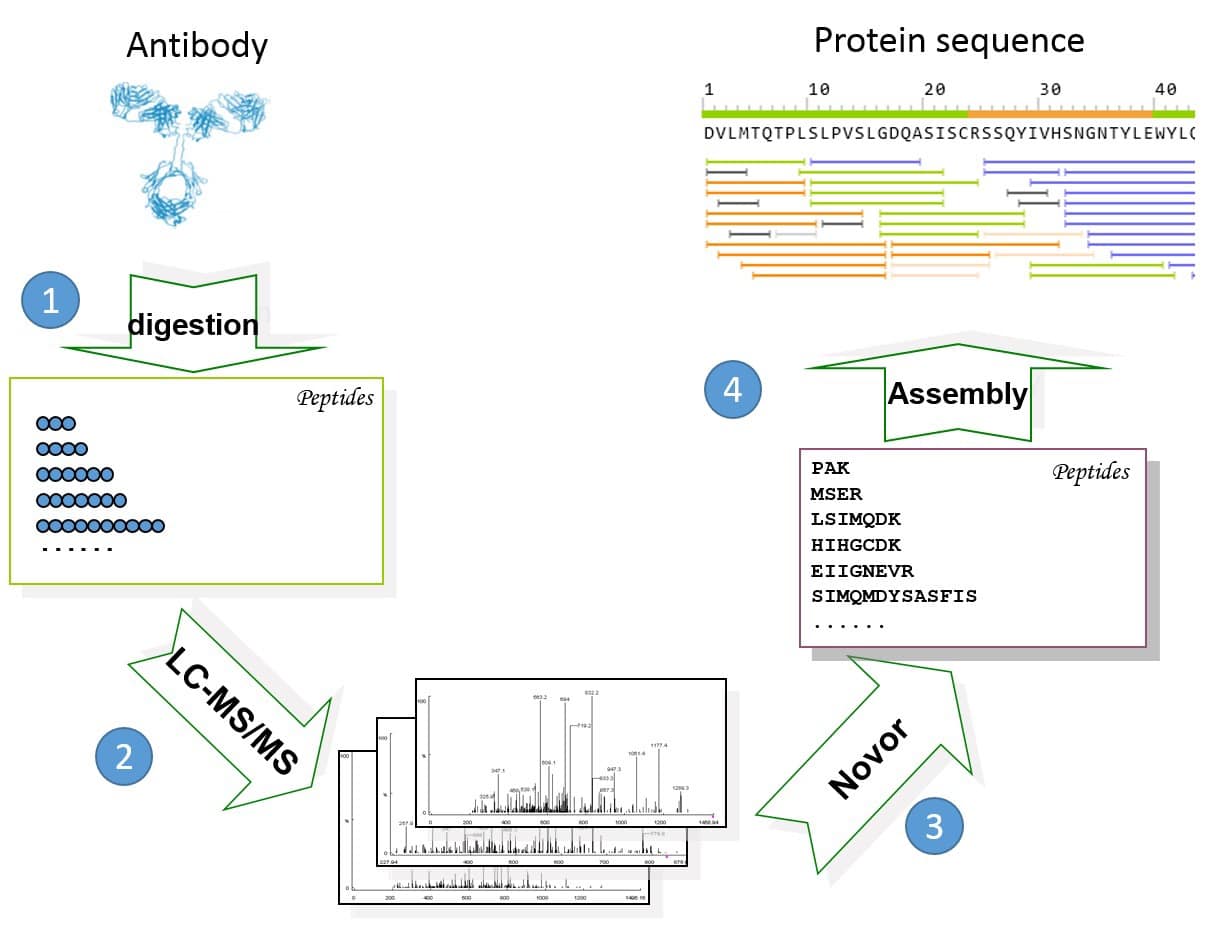
Figure 1. General workflow of REmAb® includes (1) multiple enzyme digests, (2) mass spec, (3) de novo peptide sequencing, and (4) sequence assembly.
Results
Eighty distinct research-purpose mAbs were sequenced. The majority of the antibodies are from mouse and rat, accounting for 58 and 19 of the antibodies, respectively. The other three antibodies are from three other mammals.
For all of the 80 distinct antibodies, REmAb® could confidently de novo sequence the complete sequence for each the primary pair of heavy and light chains.
In addition, a second different light chain sequence could be detected in 11 out of the 80 antibodies, or in approximately 14% of the cases. All of the 11 secondary light chains could be supported by at least ten unique peptide spectrum matches from the mass spectrometry data.
For nine of the 11 cases, the second light chain was abundant enough to get complete sequence coverage with the mass spectrometry data. For the other two cases, the MS data could confidently cover 96% and 88% of the amino acids in the variable regions, respectively.
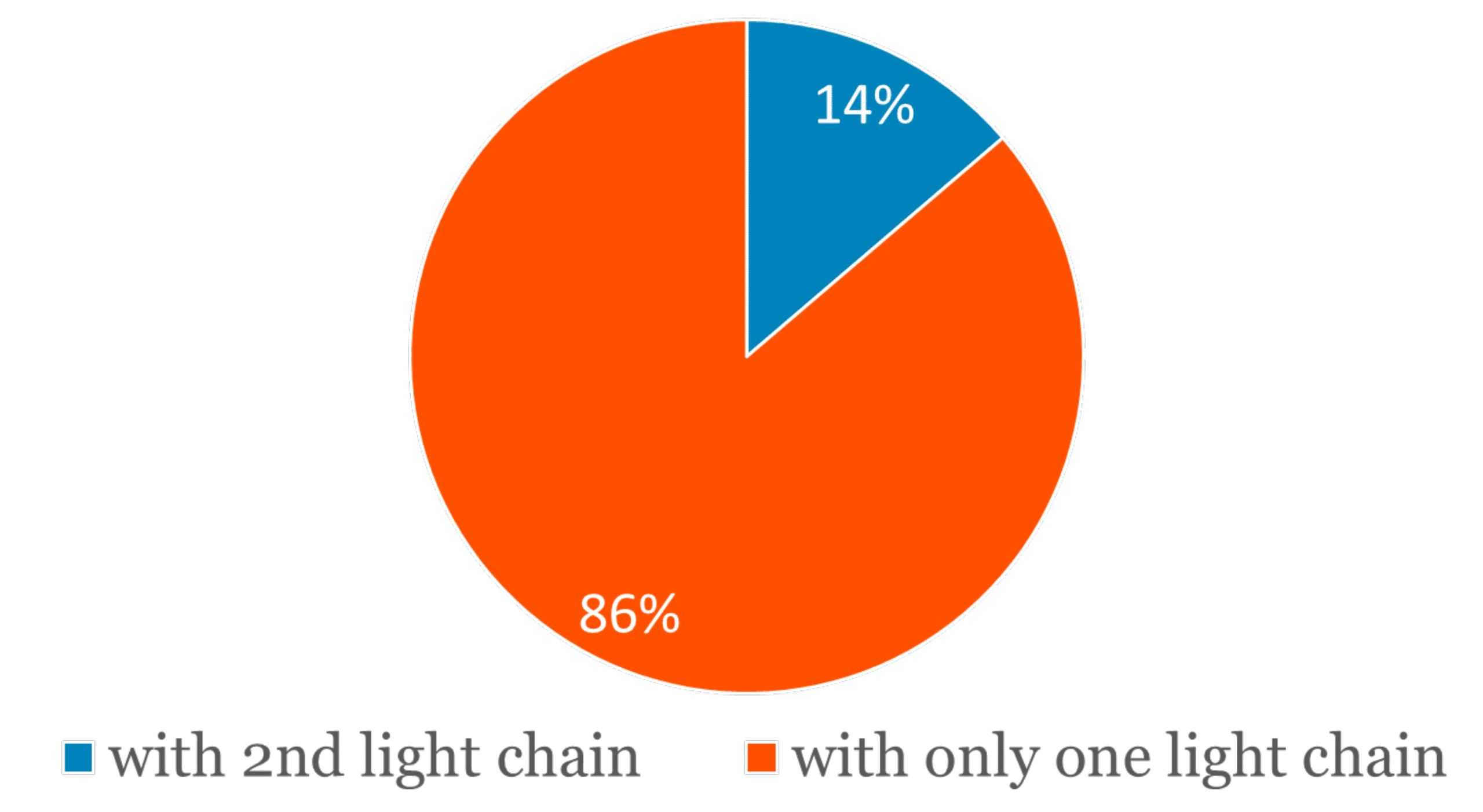
Figure 2. Approximately 14% of the studied mAb samples have an abundant secondary light chain.
Conclusions
A significant portion (14%) of the studied research purpose mAb have more than one light chain present in the sample. The mass spectrometry-based protein sequencing service, REmAb®, can sequence both the primary and secondary chains in the protein sample.
References
- Baker, M. Reproducibility crisis: Blame it on the antibodies. Nature 521, 274-276, doi:10.1038/521274a (2015).
- Bradbury, A. R. M. et al. When monoclonal antibodies are not monospecific: Hybridomas frequently express additional functional variable regions. MAbs 10, 539-546, doi:10.1080/19420862.2018.1445456 (2018).
- Ma, B. Novor: real-time peptide de novo sequencing software. J Am Soc Mass Spectrom 26, 1885-1894, doi:10.1007/s13361-015-1204-0 (2015).
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

