Paul Taylor1, Jonathan Krieger1, Qixin Liu2, Mingjie Xie2, Lian Yang2,3, Bin Ma2,3
1The Hospital for Sick Children, Toronto, ON, Canada
2Rapid Novor Inc., Waterloo, ON, Canada
3University of Waterloo, Waterloo, ON, Canada
Abstract
In this study, we conducted a large-scale statistical analysis of protein sequencing data from samples digested with multiple proteases to understand the impact of using different combinations of proteases to improve the depth of sequence coverage in the application of de novo protein sequencing. MS data for 166 monoclonal antibodies were compiled for use in this study. Each antibody protein sample was digested separately with different proteases and analyzed by LC-MS/MS. The new Pro/Ala (A/P) protease was tested to characterize NIST and MAB04HC antibody standards. De novo peptide sequencing was performed with Novor search algorithm. Protein sequences were assembled using REmAb®. The assembled mAbs demonstrate that a combination of existing proteases with orthogonal activities significantly increases confidence scores in de novo protein sequencing; however, there is a need for new proteases targeting specific amino acid(s) (a.a.) or a.a. sequences to increase antibody sequencing accuracy.
Key Takeaways
- A combination of proteases can maximize coverage during LC-MS/MS-based sequencing
- Other orthogonal approaches also contribute to increase in accuracy for de novo protein sequencing
- REmAb®’s de novo antibody sequencing service established protocol includes orthogonal and protease cocktails optimized to deliver highly accurate and full coverage protein sequences
Introduction
Monoclonal antibodies are one of the most important protein pharmaceuticals. A critical step in antibody drug development is the in-depth characterization of the protein molecule, including the primary sequence, mutations, glycosylation, and other important modifications. Multiple experiments are usually required for obtaining all such information. Human intervention is the norm for the analyses of the data from different sources. As such, the in-depth characterization of an antibody protein is currently a long and error-prone process. In this work a fully automated data analysis workflow based solely on LC-MS/MS is developed to characterize an antibody in-depth.
Materials & Methods
The monoclonal antibody protein is reduced, alkylated, and digested with six enzymes: Tryspin, Chymotrypsin, AspN, GluC, Proteinase K, and Pepsin. LC-MS/MS is performed on each digest. Novor software is used for de novo peptide sequencing. An in-house database search software, FasterDB, is used to find reference sequences from an antibody database. The de novo peptides are mapped to the reference to determine their relative positions. The consensus of the de novo sequences are taken as the real protein sequence. Once the primary sequence is determined. The MS/MS spectra are mapped to the derived protein sequence again with FasterDB for PTM and glycosylation characterization. The peak areas of each PTM and glycosylation form is calculated. Leucine and isoleucine are disambiguated by the combined use of their frequencies in the antibody database and the digestion specificity of Chymotrypsin and Pepsin.
Results
The workflow is tested with the Waters’ IgG-I antibody standard (product number 186006552). The full sequences for both the heavy and light chains are fully recovered (Figure 1) with high coverage for each amino acid.
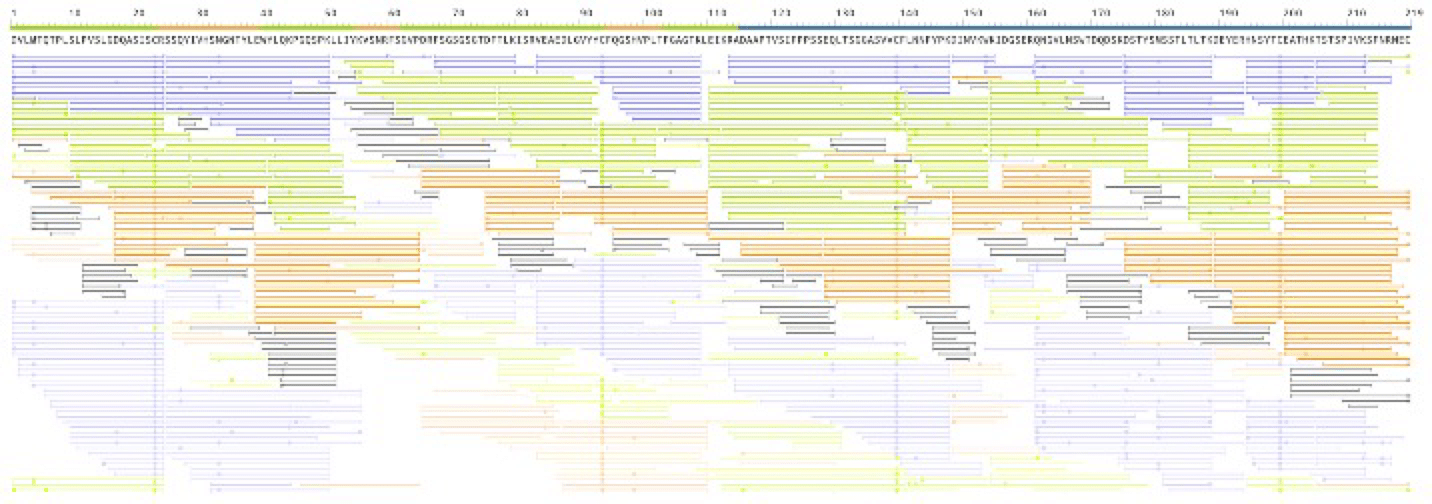
Figure 1. Software’s interactive coverage view for light chain. Each AA is covered by multiple peptides. Colors indicate peptides from different enzyme digests. Paled color indicates the peptide’s peak area < 0.1%. The ruler at the top highlights the CDR, variable, and constant regions of the antibody. Heavy chain coverage is similar.
Compared to the sequence provided by Waters, two variations were discovered on the heavy chain. The first replaces two amino acids at 49-50 from MG to GM, and the second replaces three amino acids at 68-70 from SIT to TIS. Both changes are on the variable region and are supported by strong MS/MS signal peaks (Figures 2). Since the variations do not change the intact mass of any tryptic peptide, and only slightly change the MS/MS spectra, they can only be picked up with de novo peptide sequencing. Peptide mapping or a homology-based sequencing would have failed in detecting these mutations.
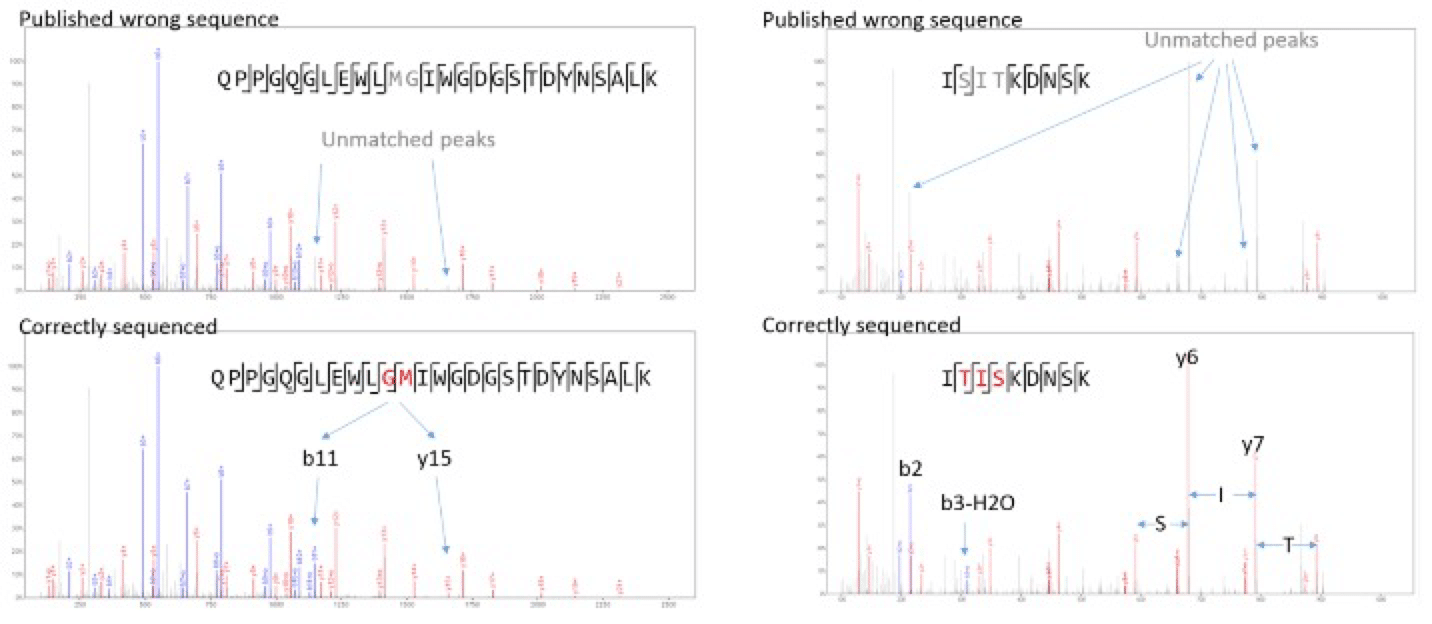
Figure 2. Evidential spectra for the mutations at heavy chain 49-50 (published MG vs our sequenced GM), and 68-70 (published SIT vs. our sequenced TIS). These mutations could not have been detected with a homology-based method as the published wrong sequences already match the spectra significantly.
Additionally, six glycosylation forms were identified (Figure 3). Five out of the six identified forms have slightly different retention time. Thus, they are not the result of in source fragmentation during MS.
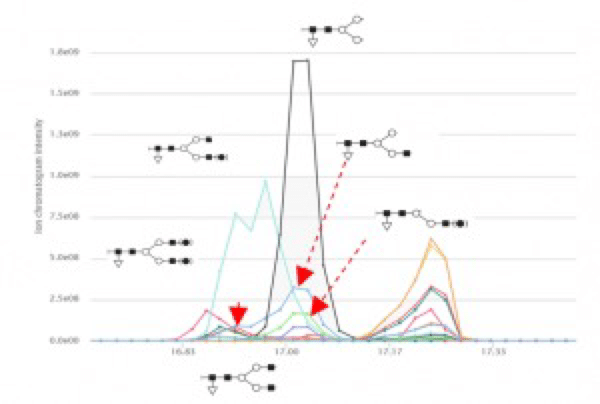
Figure 3. Glycosylation forms identified by the workflow.
Conclusions
The workflow can routinely de novo sequence the antibody proteins and profile the glycosylation. The amino acid inference is based on de novo sequencing (namely a “true de novo” method). This allows the detection of more mutations than a homology-based sequencing method.
This case study was adapted, with permission, from Taylor, P, Krieger, J, Liu, Q, Xie, M, Yang, L, Ma, B. (2016). In-Depth Characterization of Monoclonal Antibodies with a Single Experiment and Fully Automated Data Analysis. ASMS 2016 San Antonio, Texas, MP 018.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

