Written by: Genya Gorshtein, MSc
Written by: Genya Gorshtein, MSc
Published: April 3, 2024
Introduction
Membrane proteins orchestrate a wide range of cellular functions, such as cell signaling, catalysis, and ion channels. Dysregulation of these proteins frequently underlies various disease states, making them appealing targets for drug development and key players in anticancer therapies.
Identifying and effectively targeting membrane protein antigens for monoclonal antibody therapies and immunotherapies, such as antibody-drug conjugates (ADCs) and CAR-Ts is crucial for advancing next-generation therapeutics. However, this is challenging due to the nature of membrane proteins, characterized by their intrinsic hydrophobicity, complex topologies, and dynamic conformations. Overcoming these hurdles requires innovative strategies to increase the success rates of antibody discovery against membrane targets. In this discussion, we will delve into the intricacies of targeting membrane proteins and explore strategies to optimize antibody discovery success in this challenging domain.
Membrane Proteins are Difficult Antigens
Many mAb and immunotherapy drugs target transmembrane proteins (Figure 1), including programmed cell death-1 (PD-1), HER2, and cluster of differentiation (CD) receptors, such as CD19-specific chimeric antigen receptor (CAR) T cell therapies. Indeed, >60% of all approved drugs target transmembrane proteins, despite comprising only 20-30% of the proteome.
Despite their popularity as drug targets, antibody discovery and generation against transmembrane proteins remains challenging due to several factors:
- Poor expression: Membrane proteins are often expressed in low abundance on the plasma membrane. Overexpression of transmembrane proteins within mammalian cell lines may often lead to protein aggregation and cellular toxicity.
- Low solubility: Membrane proteins exhibit low solubility when removed from the phospholipid membrane due to their hydrophobic nature. This characteristic makes them challenging for in vivo antibody discovery methods, primarily due to difficulties in protein purification.
- Nonnative conformations: Due to the hydrophobic nature of membrane proteins, their native conformations need to be maintained in lipid or lipid-mimicking environments for effective antibody discovery campaigns. Extraction from the lipid bilayer may result in protein denaturation and loss of functionality.
- Limited immunogenic regions: Transmembrane proteins often present limited accessible immunogenic regions due to the majority of their structure being embedded within the lipid bilayer of the cell membrane, coupled with small extracellular loops. This characteristic complicates the process of identifying and targeting specific epitopes for antibody binding.
- Conformational changes: Membrane proteins are dynamic and undergo conformational changes throughout different binding states. Antibodies must recognize specific epitopes that may be conformational, necessitating an understanding of the protein’s conformational state for effective antigen-antibody interactions.
- Low immunogenicity and immune tolerance: Some membrane proteins exhibit low immunogenicity due to highly conserved sequences and frequent exposure to the immune system. This further complicates the process of generating an immunogenic response to obtain high-affinity and functional antibodies against these targets.
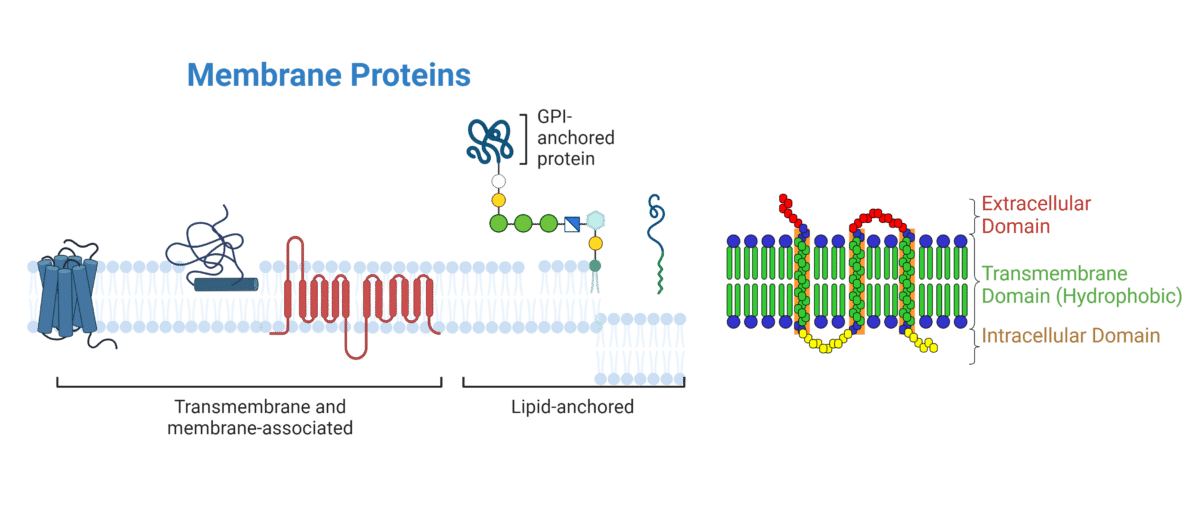
Figure 1. Types of transmembrane proteins and their domains.
Strategies for Utilizing Membrane Proteins as Antigens in Antibody Discovery
Peptides and Soluble Protein Antigens
Membrane proteins exhibit hydrophobic characteristics, requiring strategies to render them soluble for antibody discovery and generation. One effective approach involves engineering recombinant protein fragments or fusion proteins incorporating peptides derived from the native membrane protein. This method eliminates the need for producing the full-length transmembrane protein. This is difficult to do because of low expression yields, difficulty in purification, and insolubility.
The linear nature of this method limits the ability to develop conformational epitopes, and can thereby hinder the discovery of functional antibodies. Conformationally constrained peptides offer a solution by restricting the flexibility of peptides, thereby inducing specific secondary structures that can mimic the conformational epitopes of the native membrane protein (Figure 2A). For example, structured peptides derived from the G-protein coupled receptor (GPCR), CXC chemokine receptor-2 (CXCR2), have been utilized to mimic the ligand-binding site of the receptor. These mimic-peptides served as antigens in antibody discovery campaigns, resulting in the generation of functional antibodies against the desired conformational epitope. These antibodies effectively hinder ligand binding onto CXCR2 by targeting the specific conformational epitope of interest.
Identifying epitopes capable of eliciting functional antibodies with desired properties poses a significant challenge. Computational methods and in silico modeling for epitope prediction can aid in identifying immunogenic regions within membrane proteins. These computational approaches aid in selecting both linear and conformational epitopes from the transmembrane protein for further development into peptides and soluble proteins suitable for use as antigens in antibody discovery.
Whole Cells as Antigens
Whole cells can be used as a source antigens for immunization in antibody discovery campaigns (Figure 2B). However, the low abundance of membrane proteins on the plasma membrane presents a challenge and requires strategies for their overexpression.
Bacterial cells are suboptimal for overexpressing membrane proteins due to the absence of post-translational modifications and differences in protein-folding. While bacterial cells are capable of high expression levels, this does not guarantee functional and properly folded proteins.
Overexpression of human membrane proteins is most successful in mammalian cells, though it comes with challenges such as cellular toxicity, especially with proteins like ion channels. Balancing high protein expression with cell viability is crucial and can be regulated using techniques like gene expression control through an inducing system. Additionally, monitoring overexpression in mammalian cells faces challenges due to the lack of available antibody reagents. Epitope tags and anti-tag-specific antibodies can help mitigate this issue. For more information on choosing the appropriate expression system, check out our blog on recombinant protein expression.
Immunizations for antibody discovery often utilize Chinese hamster ovarian (CHO) cells or HEK cells as hosts for transgene overexpression. These systems are advantageous due to their availability and well-studied nature as overexpression systems, presenting the target antigen in its native conformation for optimal chances of functional antibody binding. However, the cell’s entire proteome is considered non-self by the host’s immune system. This can act as an advantage, potentially boosting the immune response, but it also means that the cell’s proteins will compete with the target overexpressed antigen for antibody binding, posing a disadvantage.
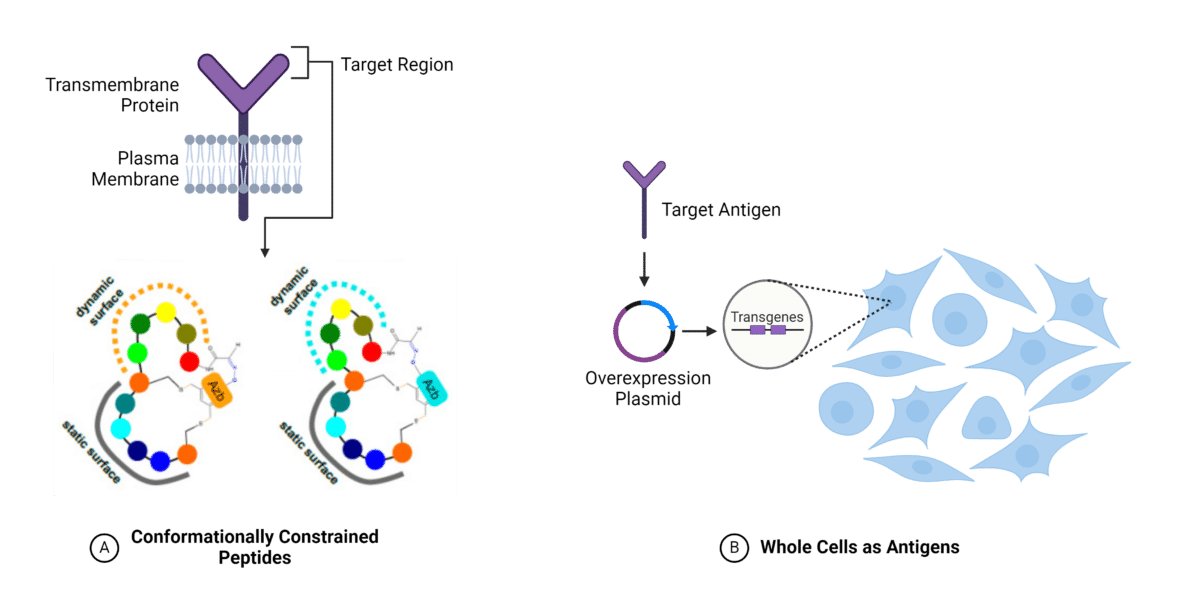
Figure 2. Methods for utilizing membrane proteins as antigens for antibody discovery. A. Conformationally constrained peptide approach. B. Whole cells as antigens with protein overexpression. (Adapted from Bozovicar et al., 2021)
Purified Protein Formats
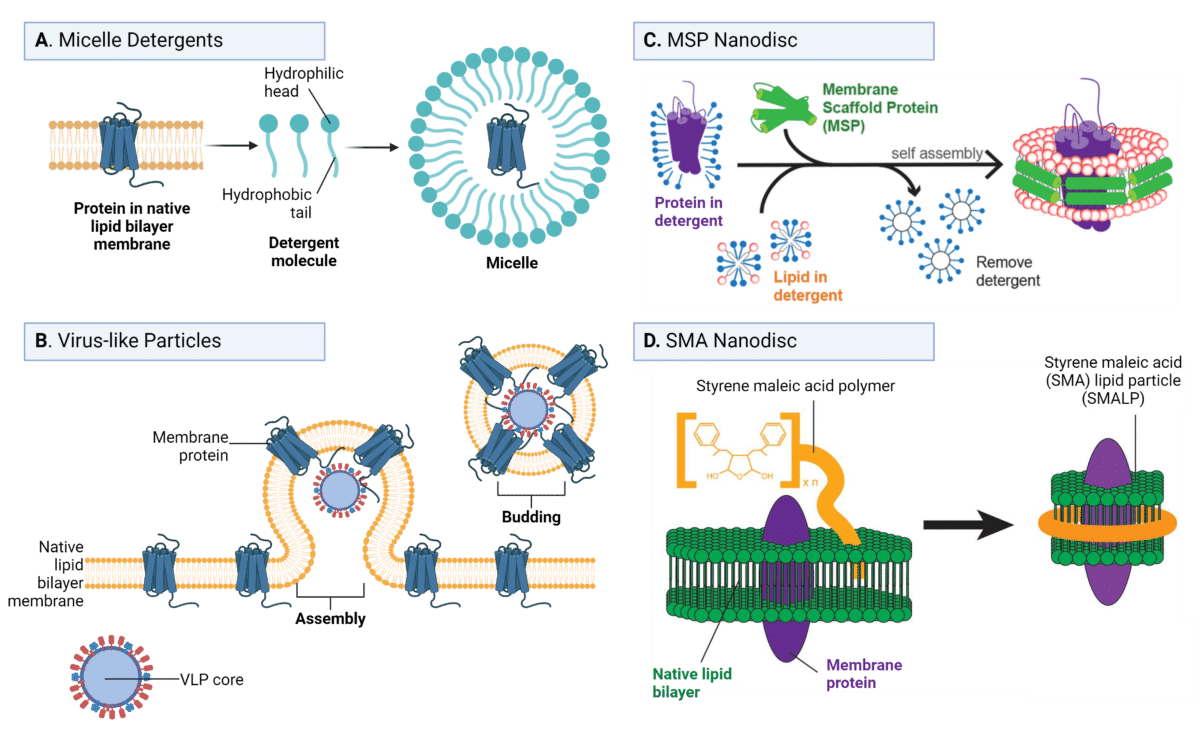
Purified proteins serve as the optimal input for antibody discovery. Nevertheless, the inherent properties of membrane proteins pose challenges to their purification. These properties necessitate strategies for their extraction from membranes while masking their hydrophobic surfaces (Figure 3).
Micelle Detergents
Traditionally, detergents have been employed for this purpose, as they are amphipathic molecules comprising hydrophilic heads and hydrophobic tails. These detergents aggregate to form micelles, enveloping the hydrophobic membrane proteins and facilitating their extraction from the membrane while maintaining stability (Figure 3A).
Despite significant advancements in membrane protein purification using detergent micelles, challenges persist in their application for antibody discovery:
- Hydrophobic tails may obstruct antibody binding to extracellular loops, thereby impeding detection.
- Detergents can interfere nonspecifically with antibody binding in certain discovery modalities, such as phage display.
- Disassociation of detergents from the micelle can lead to denaturation and the loss of conformational epitopes, compromising antibody recognition.
Additional cell-free antibody engineering techniques may be employed to manipulate glycans, including modifications to amino acid sequences for the removal of glycosylated residues or Fc-swapping to incorporate Fc domains with desired glycan modifications and effector functions.
Virus-like Particles
Virus-like particles (VLPs) offer a lipid bilayer-based antigen method capable of preserving native protein formats (Figure 3B). These self-assembled structures consist of capsid proteins that closely mimic the size and structure of a virus particle, yet lack genetic material, rendering them incapable of autonomous replication or infecting host cells. VLPs can be expressed in various systems, including bacteria, mammalian cells, and cell-free systems, providing flexibility in production.
VLPs induce plasma membrane budding and subsequent release, leading to the isolation of the target membrane protein from the cell within its native phospholipid bilayer, which can then be purified and used for immunization in antibody discovery campaigns.
Advantages of VLPs in maintaining native protein formats include:
- Preservation of Native Lipid Bilayer: VLPs maintain a native lipid bilayer environment, thereby keeping additional membrane components intact, such as sphingolipids. This preservation ensures the inclusion of relevant lipid species that may be crucial for the proper folding, stability, and function of the membrane protein.
- Maintenance of Correct Membrane Protein Orientation: VLPs ensure the correct orientation of membrane proteins, with their extracellular domains exposed to antibody binding and the intracellular domains hidden. This orientation is critical for eliciting specific antibody responses against extracellular epitopes, which are often the targets of interest in antibody discovery campaigns.
Lipid Nanodiscs
As an alternative to employing detergents, the nanodisc technique facilitates the utilization of membrane proteins as antigens in more native-like formats. This method involves embedding the membrane protein into a lipid bilayer disc, with the hydrophobic edge of the disc encircled by two copies of an amphipathic apolipoprotein A1 derivative, known as a membrane scaffold protein (MSP) (Figure 3C). However, embedding the membrane protein within the MSP-nanodisc necessitates the use of detergents, thereby introducing risks to stability and activity.
Another alternative disc format is the styrene maleic acid (SMA) lipid particle (SMALP), wherein MSP is replaced by a cost-effective amphipathic SMA polymer (Figure 3D). SMA can readily extract membrane proteins directly from the lipid bilayer without the need for detergents.
Compared to MSP nanodiscs, SMA nanodiscs offer several advantages:
- Preserve the native protein structure and function by circumventing the use of detergents.
- SMA serves as a non-protein scaffold, mitigating the generation of anti-scaffold antibodies.
- Conjugation of SMA to fluorophores or biotin is feasible, offering potential advantages for antibody discovery applications.
Figure 3. Purified protein approaches for using membrane proteins as antigens for antibody discovery. A. Micelle detergents, B. Virus-like particles, C. MSP nanodisc, and D. SMA nanodisc. (Adapted from Yeh et al., 2018 and de Jonge et al., 2020).
Antibody Discovery Services
Membrane proteins pose challenges as antigens, yet they continue to be the primary targets for novel monoclonal antibodies (mAbs) and immunotherapies. Despite these challenges, numerous strategies can enhance the success of generating unique and functional antibodies against these targets. The selection of a strategy depends on the antigen’s specific requirements, the chosen antibody discovery platform, and the intended application.
Rapid Novor’s antibody discovery service using mass spectrometry enables the generation of unique and functional antibody binders against difficult antigens. REpAb® is species agnostic, allowing for a wide variety of hosts for antibody discovery, and it is adaptable to various antigen formats, including peptides, proteins, enzymes, and small molecules.
Reach out to our scientists to discuss antigen development, immunization, and antibody discovery tailored to your challenging antigens.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics