 Written by: Genya Gorshtein, MSc
Written by: Genya Gorshtein, MSc
Published: May 29, 2024
Introduction
An epitope is an inherent property of an antibody that cannot be altered without creating an entirely new antibody, making it arguably the most critical factor in determining antibody functionality.
Epitope binning, an important early-stage screening process, involves testing antibodies pairwise in a competition immunoassay to identify which antibodies block the same epitopes on a target antigen. Surface plasmon resonance assay has emerged as a powerful analytical tool for this purpose, offering a label-free, high-throughput method for binning antibodies. Implementing epitope binning in the target-to-hit stages of antibody drug discovery enables early characterization and selection of functional candidates for downstream engineering and optimization.
What is Epitope Binning
Epitope binning is the testing of antibodies in a pairwise, competition immunoassay to identify which antibodies bind to overlapping or adjacent epitopes on the target antigen. A blocking profile is established for each antibody, and antibodies with similar blocking profiles are grouped together in a common bin process known as “epitope binning”.
While the criteria for grouping antibodies in the same bin can vary, they are all considered to be competing for the same epitope. In contrast, antibodies from different bins are considered to be compatible for applications that require discrete epitopes.
The main goal of epitope binning is to classify antibodies based on their binding profiles and identify groups that recognize distinct or overlapping epitopes, thereby revealing their functionality.
Epitope Binning Assay Set Up with SPR
In contrast to other methods like conventional ELISAs, which have limited dynamic range, lower specificity, require large sample volumes, and involve labeling steps, surface plasmon resonance (SPR) stands out as the gold standard for epitope binning. SPR can provide real-time, label-free, continuous monitoring and detection of diverse biomolecular interactions, suitable for complex biologics such as bispecifics, multispecifics, nanobodies, and antibody-drug conjugates (ADCs). We have previously discussed the underlying theory of SPR and its application for characterizing biomolecular interactions.
Binning Formats
The 3 main binning assay formats include: sandwich, premix, and tandem approaches (Figure 1).
- Sandwich binning assay: The first mAb is covalently attached to the surface of the sensor. The antigen is then injected and binds to the first mAb, followed by injection of the second mAb. If the second mAb binds to the antigen, it creates a ‘sandwich’, indicating that they have different epitopes and belong to separate bins. Alternatively, if the second mAb is blocked from binding to the antigen by the first mAb, it suggests that they have overlapping epitopes and should be binned together.
- Premix assay: Similar to the sandwich assay format, the first mAb is covalently attached to the biosensor. The antigen and the second mAb are pre-mixed outside the instrument at saturating conditions and then injected as a complex.
- Tandem assay: In the tandem binning assay, the antigen is covalently attached to the sensor surface. The first mAb is injected under saturating conditions, followed by injection of the second mAb. If both mAbs bind to the immobilized antigen, it indicates that they have different epitopes and belong in different bins. Conversely, if the second mAb does not bind to the antigen, it suggests that the first mAb is blocking the epitope, binning them together because of overlapping epitopes.
Each assay format has its advantages and disadvantages. As such, choosing an assay format is dependent on the antibody and antigen being investigated. For example, the tandem assay format is well-suited for more complex, multivalent antigen targets and requires low amounts of antigen for immobilization.
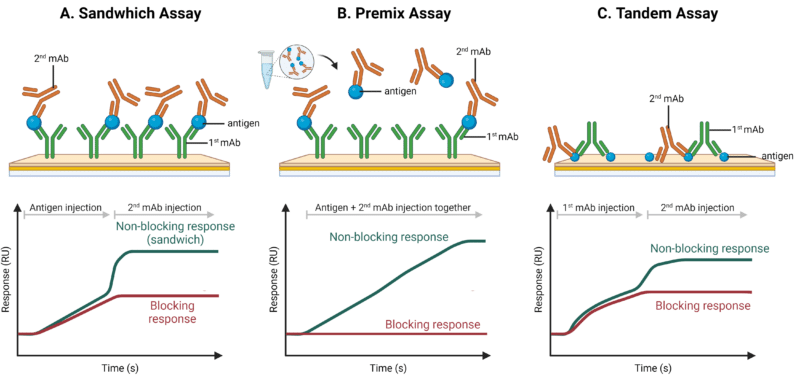
Figure 1. Schematic representation of 3 different binning format set ups and their sensorgrams. A. Sandwich assay, B. Premix assay, and C. Tandem assay.
Symmetrical vs. Asymmetrical Binning Assays
Epitope binning is a pairwise analysis that can be conducted in symmetrical or asymmetrical formats. In an asymmetrical binning assay, each antibody is tested individually against a panel of other antibodies. In symmetrical binning assays, each antibody is tested against every other antibody by reversing the order in which the antibodies bind to their antigen. As such, a 4 antibody binning assay involves 16 interactions, a 10 antibody binning analysis involves 100 interactions, binning 30 antibodies involves 900 interactions, and so on.
Symmetrical epitope binning assays are preferable because the sequence in which antibodies are injected can influence binding or blocking results. For instance, in a 4×4 tandem binning experiment of alpaca mAbs, different antibody functions were observed depending on their role as the first saturating antibody versus the second analyte antibody (Figure 2).
In this experiment, 4 clones (R1, R4, R7, and R9) were binned against the P17 antigen to elucidate epitope diversity. When R4 was used as the saturating antibody, it showed low-level decreases in binding response (0-27% blocking) against other clones. However, when R1, R7, and R9 were used as saturating antibodies, they significantly reduced R4’s ability to bind to the antigen (56%-71% blocking).
Directionality of blocking is often encountered in binning assays, emphasizing the importance of the binding sequence even between antibodies from separate bins.
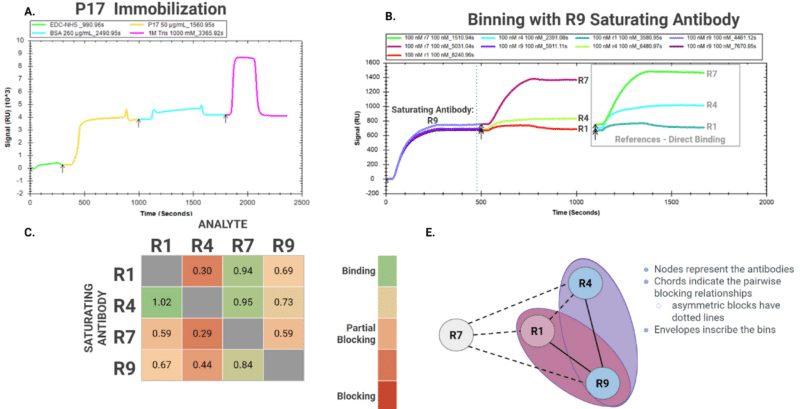
Figure 2. Example of a 4×4 tandem binning assay for alpaca mAbs (R1, R4, R7, R9) against antigen P17. Each of the clones was injected at a saturating 100 nM concentration and used either as a saturating antibody or a binding antibody. A. Sensogram for antigen immobilization. B. Sensogram for epitope binning experiments. Here, R9 is the first saturating antibody, and R1, R4, and R7 were injected at the second mAbs across three different experiments. C. Heat-map of the results of the 16 binning experiments. Green indicates a binding relationship, while red indicates a blocking relationship. Values indicate the degree of binding. D. Illustrates the bins and epitope relationships among the four clones.
Value of Epitope Binning in Early Discovery Stages
Epitope binning is essential in the early discovery stages of antibody therapeutic pipelines, as it addresses key limitations of traditional affinity selection methods. Typically, mAbs with the highest affinity are chosen for further study, but this approach can restrict candidates to a small number of epitopes (Figure 3). If these epitopes are not functionally relevant for the intended therapeutic use, it can result in a “dead-end” with no viable alternatives. These dead-ends may be detected during early functional tests, but they often appear later in the drug development process, leading to significant delays and increased costs.
Utilizing epitope binning allows researchers to test top antibodies from each bin, facilitating the quicker and more cost-effective identification of therapeutically relevant epitopes. This method provides important information earlier in the discovery phase, reducing cycle time and overall expenses, and increasing the likelihood of developing successful antibody therapeutics.
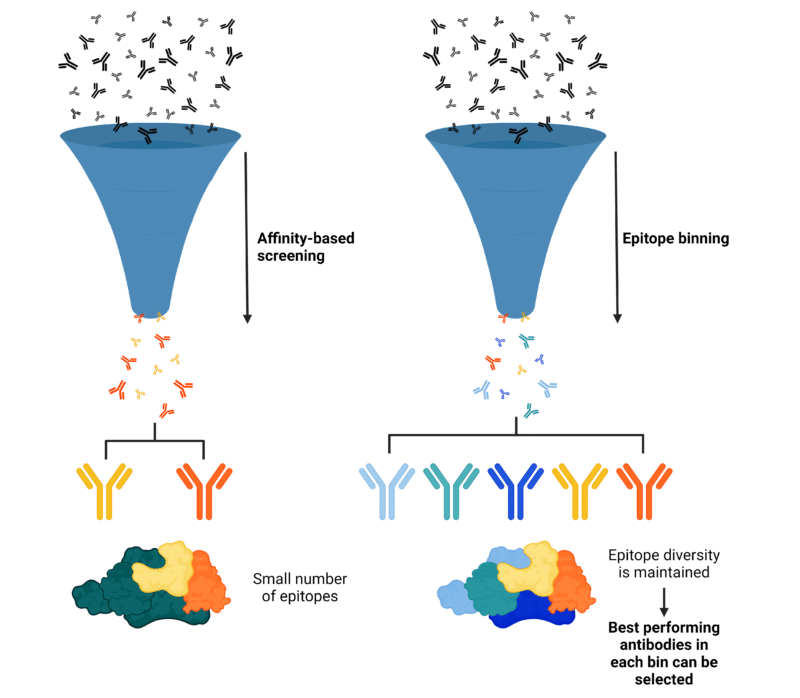
Figure 3. Illustration of antibody development pipeline and outcomes with affinity-based screening approaches vs. epitope binning is conducted during early stages of development. Affinity-based screening (Left) can be biased towards few epitopes, which limits the probability of identifying functional epitopes and can miss candidates with desirable qualities. When epitope binning (Right) is employed in early development stages, epitope diversity is maintained, providing information on antibody functionality, allowing for selection of the best performing antibodies from each bin for further characterization, engineering, and optimization.
Epitope Binning with SPR on Biacore 1K
Our SPR service uses label-free evaluation of epitope binning, utilizing surface plasmon resonance (SPR) analysis technology from Biacore. This nanotechnology-based SPR assay facilitates the immobilization of binding partners in both orientations on the sensor chip, ensuring minimal bulk shift, high sensitivity, and low noise, while maintaining high throughput.
In the case of a large number of candidates selected for screening, practical considerations necessitate the use of complementary techniques. Methods such as SPR kinetic screening and in silico AI-based epitope mapping can help focus the binning experiment on a smaller number of antibodies.
Understanding of antigen-antibody interactions also requires analysis into 3D structures to fully elucidate the binding epitopes of antibodies. Our epitope mapping service can identify the binding site of an antibody to its corresponding antigen with high resolution, down to 1-5 amino acids.
Contact our scientists to learn more.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

