Authors:
Teresa Nunez, Amber Couzens, Jin Duan, Chelsea Reitzel, Rosalin Dubois,
Lin Wu, Qixin Liu, Thierry Le Bihan, Marko Jović, Dominic Narang, Bin Ma
Published: Nov 13, 2023
Abstract
In this study, the challenge of accessing functional antibodies from the circulating antibody repertoire was addressed using Rapid Novor’s antibody discovery platform, REpAb®. By employing polyclonal antibody sequencing technology using mass spectrometry, we sequenced and discovered functional antibodies directly from immunized alpaca serum. These antibodies were further characterized using techniques such as Hydrogen-Deuterium Exchange Mass Spectrometry (HDX-MS) and Surface Plasmon Resonance (SPR) to elucidate their binding epitopes and kinetics. The integration of polyclonal antibody sequencing with HDX-MS and SPR provided a comprehensive workflow, enabling the discovery and detailed characterization of biologically relevant antibodies.
Key Takeaways
- Sequencing of the alpaca IgG proteome de novo is capable of identifying the physiologically relevant, high affinity binders while not being limited by the availability of B-cells in circulation.
- SPR was employed for kinetic analysis and epitope binning of antibody candidates to investigate the binding behaviors and epitope diversity between the antigen and candidate mAbs.
- HDX-MS was used to identify binding epitopes in different regions of the rabbit IgG antigen, demonstrating the epitope diversity of these recombinant alpaca mAbs, potentially indicating different targets and mechanisms of action.
- Combined approach between machine-learning driven proteomics, SPR and HDX-MS makes it possible to accelerate the antibody discovery workflow and lead selection by minimizing the need to produce the entire collection of potential candidate antibodies.
Introduction
The circulating antibody repertoire contains a rich pool of biologically relevant antibodies. Accessing these antibodies, whose B-cells underwent several rounds of in vivo affinity maturation to yield dozens of high-affinity binders, has been an unaddressed technological challenge in antibody discovery. Interrogating the antibody repertoire through peripheral blood samples is the only observable interface between germinal centers and bone marrow, and has the potential to discover the most biologically and functionally relevant antibodies of the repertoire.
To overcome this obstacle, Rapid Novor pioneered the development of REpAb®, an innovative antibody discovery platform. REpAb® utilizes polyclonal antibody sequencing technology with mass spectrometry to directly sequence monoclonal antibodies (mAbs) from serum samples (Figure 1). We sequenced and discovered functional antibodies directly from immunized alpaca serum raised against a large rabbit IgG antigen. We recombinantly expressed the antibodies and subjected them to in-depth characterization with Hydrogen-Deuterium Exchange Mass Spectrometry (HDX-MS) and Surface Plasmon Resonance (SPR) to elucidate the binding epitopes and kinetics.
Here we demonstrate the application of REpAb® polyclonal antibody sequencing coupled with HDX-MS and SPR to provide an integrated workflow from antibody discovery to lead characterization and selection.
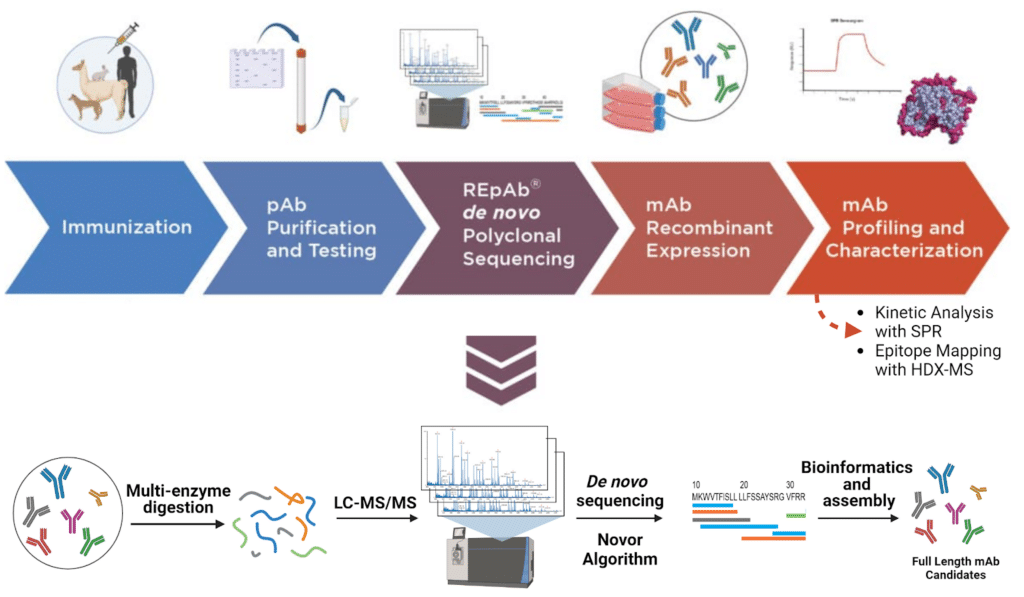
Figure 1. Antibody discovery and characterization pipeline
Methods
De Novo Sequencing
Immunized alpaca serum was digested with several different protease enzymes, and digests were analyzed by liquid chromatography with tandem mass spectrometry using an Orbitrap Eclipse Series Instrument (Thermofisher Scientific, CA, US) coupled to the LC Evosep One (Evosep, Denmark). De novo peptide sequencing was performed with proprietary software that is an evolution of the Novor search algorithm, and the protein sequencing was assembled with REpAb® using machine learning (ML)-based bioinformatics.
Epitope Mapping with Hydrogen-Deuterium Exchange Mass Spectrometry (HDX-MS)
For HDX-MS analysis, antigen digestion was performed using the pepsin/protease XIII column. Peptides were identified using the Waters™ SELECT SERIES Cyclic IMS mass spectrometer.
Kinetic Analysis and Epitope Binning with Surface Plasmon Resonance (SPR)
Kinetic analysis of recombinant mAbs was performed using openSPR by Nicoya with a carboxyl sensor.
Results and Discussion
10 Unique mAb Sequences Were Discovered from the Polyclonal Mixture
Using our mass spectrometry (MS)-based REpAb® antibody discovery platform combined with machine learning (ML)-based bioinformatics, IgG mAbs were sequenced de novo directly from the alpaca immunoserum (Figure 2A). Ten unique full length alpaca mAb sequences were discovered from the polyclonal mixture and expressed recombinantly. All 10 recombinant mAbs demonstrated affinity towards the rabbit IgG antigen (Figure 2B).
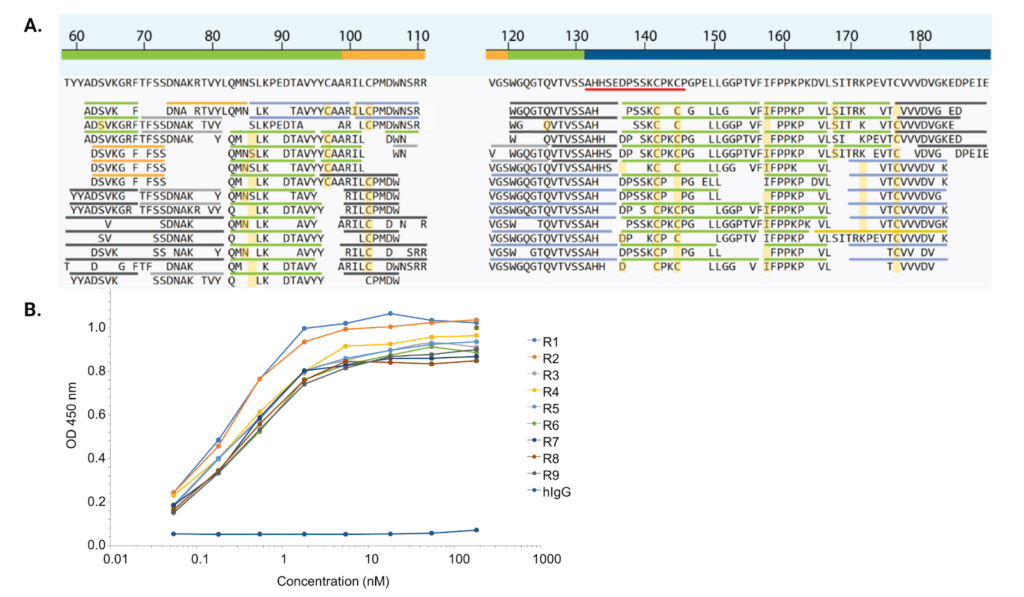
Figure 2. De novo sequencing and recombinant ELISA results. A. De novo sequencing example of a natural sequencing found in a polyclonal mixture. Labelled according to the CHOTIA scheme , the top line is the region assignment of the sequence, with green representing the framework region, orange representing the CDR3, and blue representing the constant region. Below the region assignment is the sequence consensus, and the underlined sequence beginning with AHHSEDPSSKCPKCP is the hinge region specific to IgG2c. Additionally, the shorter sequences below represent the different digests performed on the mixture, with green representing chymotrypsin, blue representing trypsin, light orange representing lysC, dark orange representing AspN, and black representing pepsin. Each amino acid is represented by more than 30 occurrences, with only the top 13 most intense peptides per region being illustrated. B. ELISA analysis of recombinant alpaca antibodies expressed with human Fc. All recombinantly expressed antibodies show affinities in the low nM range.
Kinetic Analysis with SPR Reveals Four High Affinity Binders
Of the ten mAb sequences isolated from the alpaca immunoserum, four were selected for downstream characterization. Using SPR, full kinetic profiles (Kon, Koff, and KD) were determined for R1, R4, R7, and R9 against 2 rabbit mAb antigens (P17 and CD3e) (Figure 3A). Each of the four binders exhibited a unique binding profile with a potential for further downstream antibody development in a variety of applications. Key defining characteristics of the set are the slow on-rate of the R7 clone (0.63×105 M-1 s-1) and the fast off-rate of the R1 clone (61.9×10-4 s-1) (Figure 3B).
R4 and R9 clones were found to have similar kinetic profiles with high affinity towards both P17 and CD3e mAb targets, with a slight preference for CD3e mAb. The primary driver of the lower affinity of R1 is the off-rate that is an order of magnitude faster than that of R4, R7 and R9. Finally, the weaker affinity of R7 towards P17 is primarily due to the slower on-rate in comparison to all the other alpaca mAb clones.
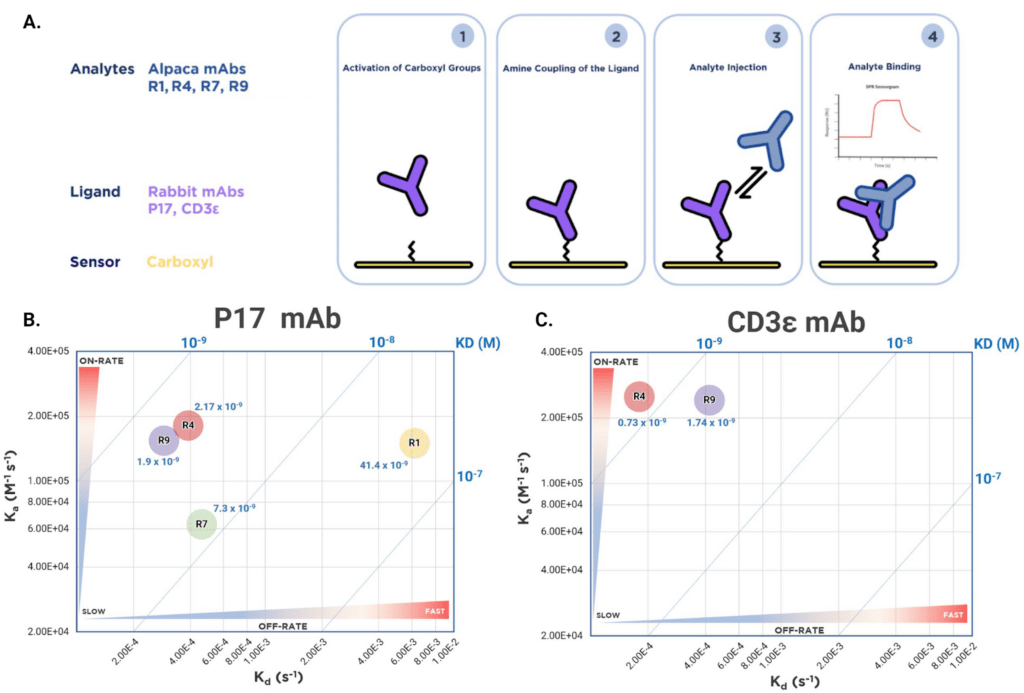
Figure 3. Kinetic analysis experimental set up and iso-affinity results of four recombinant alpaca mAbs. A. SPR experimental set-up for kinetic analysis. B. Comparative affinity analysis was performed to dissect individual contributions of each kinetic parameter for each alpaca mAb. Two-dimensional iso-affinity kinetic plot of rate constants (Kd and Ka) with blue diagonal lines depicting equilibrium binding constants based on rate constant ratios.
Epitope Binning of Four Alpaca mAbs on P17 Antigen
To investigate epitope diversity using SPR, we conducted a 4×4 tandem epitope binning experiment, examining the epitope distribution of four alpaca mAbs against the P17 rabbit monoclonal antigen. For each replicate, P17 was immobilized under identical conditions followed by an injection sequence of a blocking antibody at a saturating concentration and binding antibody serving as an analyte (Figure 4A). Tandem epitope binning was performed in both sequences, with each antibody being used as a saturating as well as binding antibody. Saturation efficiency was assessed by performing blocking and binding with the same antibody.
When being used as a saturating antibody, R4 showed the highest range of compatibility, with three other clones exhibiting relatively low level decrease in binding response (0-27% blocking). Conversely, blocking with R7 dramatically reduced epitope accessibility (41-71% blocking, Figure 4B). To understand the antibody and antigen interactions on a structural and 3D level, HDX-MS analysis was performed.
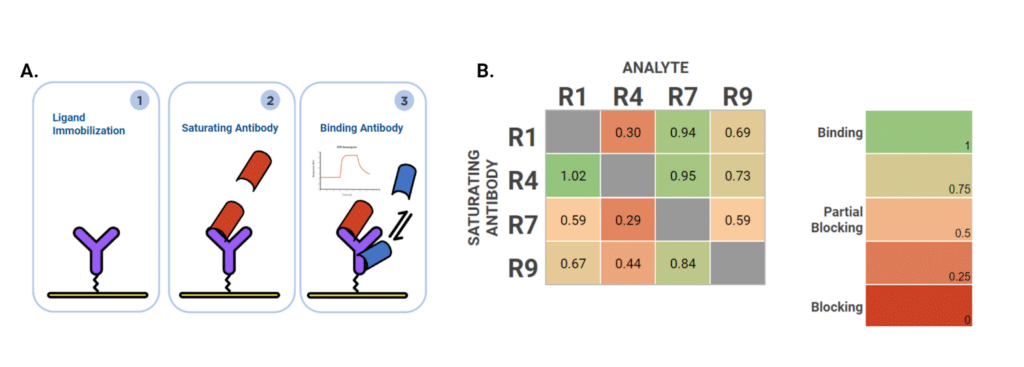
Figure 4. A. Experimental set-up for epitope binning. B. The epitope diversity on P17 was evaluated for R1, R4, R7 and R9 using a tandem binning approach. Each of the clones was injected at a saturating 100 nM concentration and used either as a saturating antibody or a binding antibody in a 4×4 binning format.
Epitope Mapping with HDX-MS Identified Four Unique Binding Epitopes on the Antigen
Using HDX-MS, we identified the epitopes of the four recombinant alpaca mAbs R1, R4, R7, and R9 on the rabbit P17 IgG antigen. Notably, each alpaca mAb exhibited distinct binding regions on the antigen, as evidenced by observable changes in deuterium fractional uptake differences between the free and bound states of the antigen, as illustrated in the heatmaps (Figure 5A-D). Analysis of R1 binding to the antigen resulted in an increase in deuterium uptake, indicated by the red on the heat map. This is unusual compared to the other 3 mAbs, where only deuterium loss (blue) was observed.
Together, HDX-MS results of mAb binding to the P17 rabbit mAb antigen reveals that R1 and R7 bind to the Fab region of the antigen, whereas R4 and R9 bind to the Fc region of the antigen (Figure 5E).
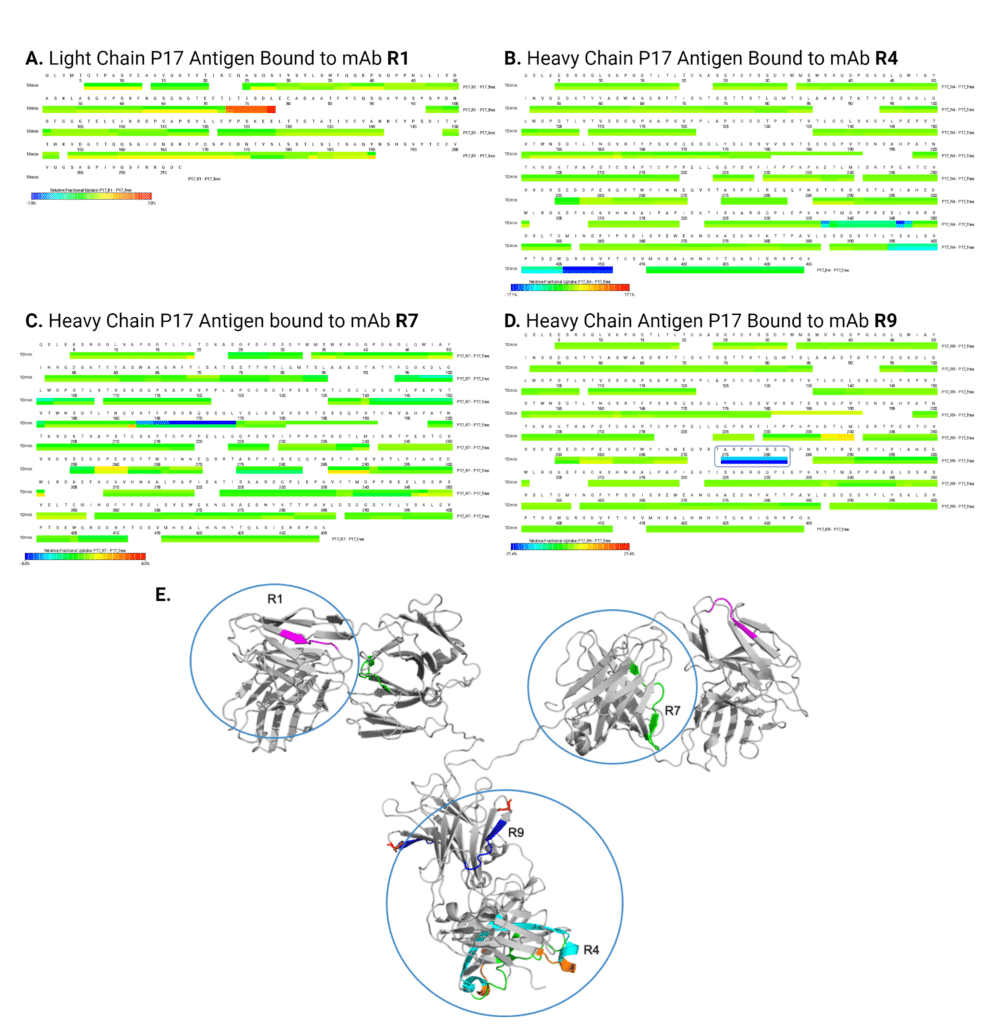
Figure 5. Results of Epitope Mapping of R1, R4, R7, and R9 against P17 rabbit IgG antigen using HDX-MS. Protein level overview of deuterium uptake of the antigen comparing bound and unbound states with: (A) LC antigen with mAb R1, (B) HC antigen with mAb R4, (C) HC antigen with mAb R7, and (D) HC antigen with mAb R9. Colours represent relative functional uptake of deuterium: blue: less uptake; green: no change in uptake; red: increased uptake. (E) An antigen homology model (PyMOL) of rabbit IgG (grey) with all different recombinant mAb epitopes mapped onto a single structure (R1, R4, R7, R9), showcasing their differences in deuterium uptake and the binding diversity of these alpaca IgG mAbs. R labels correspond to the name of each mAb.
Conclusions
This is the first successful study to sequence and characterize the alpaca IgG proteome de novo, with a technology that directly sequenced IgGs from the immune repertoire, thus identifying more physiologically relevant, high affinity binders while not being limited by the availability of B-cells in circulation. Furthermore, this study demonstrates the application of de novo polyclonal sequencing for antibody discovery, coupled with SPR and HDX-MS characterization to aid in lead identification and selection.
This study focused on Alpacas, a species that produces a unique subclass of camelid antibodies lacking light chains. These antibodies are a structurally unique family of immunoglobulins (IgGs) that carry a significant potential for biologics development and engineering given their small size, stability and high affinity. Moreover, it demonstrates that the REpAb antibody discovery approach is adaptable to various species lacking readily available B cell and genomic information. This methodology is applicable to exploring unique and novel antibody formats, showcasing its versatility and broad applicability.
More Information on Antibody Discovery and Characterization
Antibody Discovery Services
Mass spectrometry based in vivo antibody discovery platform, that sequences functional antibodies directly from serum or purified protein samples.
Explore Antibody Discovery Services
SPR Kinetic Analysis and Epitope Binning
Our Surface Plasmon Resonance (SPR) service can determine the on-rate (Kon), off-rate (Koff), dissociation constant (Kd), and affinity (KD) for antibody-antigen interaction.
Explore SPR Services
HDX-MS Epitope Mapping
Mass spectrometry based epitope mapping service, that can identify the binding site of an antibody to its corresponding antigen with the highest confidence and resolution.
Explore Epitope Mapping Services
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

