 Written by:
Written by:
Genya Gorshtein, MSc
Published: September 12, 2023
Contents
Introduction
The concept of “developability” describes the likelihood of a candidate antibody being both manufacturable and a safe and effective therapeutic drug. It is crucial to assess the developability of a candidate antibody drug early and often in the discovery stages to prevent unsuitable candidates from advancing to pre-clinical stages.
A significant concern in the developability of antibody-based drugs is related to protein stability and aggregation. Protein stability refers to a protein or antibody’s ability to maintain its native folded structure and resist denaturation or degradation. When the protein/antibody undergoes partial denaturation or degradation due to stress, it can form clumps or aggregates that can adversely impact their function, safety, and efficacy.
To address developability challenges associated with aggregate-prone and unstable antibodies, sequence analysis and antibody engineering offers promising solutions. In this discussion, we will delve into the mechanisms that impact antibody aggregation and stability and explore strategies to improve their developability.
Mechanisms That Influence Protein Aggregation and Stability
Protein aggregation is a complex phenomenon where proteins undergo conformational changes, misfolding, and self-association to create large aggregates or clumps that can impact their safety and effectiveness as biotherapeutics. The process of protein aggregation can be divided into 5 main stages (Figure 1):
- Partial unfolding of the native monomer, transitioning to its non-native conformation.
- Reversible self-association of the partially unfolded protein.
- Irreversible aggregation creates the nuclei, which is the smallest stable aggregate.
- Further growth of aggregates due to the addition of monomers.
- Addition of a large number of monomers to the aggregate results in high molecular weight, irreversible, and insoluble aggregates that precipitate out of solution.
Protein aggregation is often associated with decreased stability, but some proteins may also aggregate under their native-like conformations. However, antibodies are most susceptible to non-native and insoluble aggregation.
Several conditions can promote the aggregation of antibody, including:
- Temperature: High temperatures can cause antibody denaturation and unfolding, as well as increase the rate of aggregation. Low temperatures can cause protein denaturation and promote aggregation through freeze-thaw cycles.
- pH: Proteins and antibodies are stable at narrow pH ranges and may denature and aggregate outside of those ranges. Solution pH impacts the total protein charge, thereby affecting electrostatic interactions and can induce protein self-association and aggregate formation.
- Concentration: High protein concentrations increase the viscosity of the solution and increase aggregation potential by increasing protein-protein interactions and self-association.
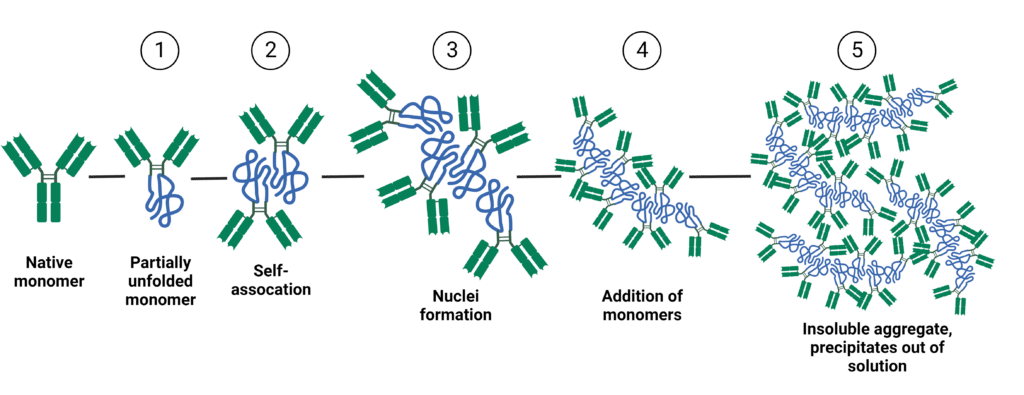
Figure 1. The 5 main stages of antibody aggregation.
Sequence and Structural Characteristics That Impact Antibody Aggregation and Stability
Antibodies are prone to aggregation due to their large hydrodynamic radii, non-symmetrical hydrophobicity, and charge distributions. Antibodies generally have multiple “hot-spots” or aggregation-prone regions (APRs), and when partial unfolding occurs, exposed regions facilitate strong intermolecular interactions that lead to aggregation.
The aggregation and instability of antibodies is not only impacted by solution conditions as described earlier, but also by intrinsic properties of the antibodies. These intrinsic factors include the primary amino acid sequences, structural motifs, and post-translational modifications (PTMs).
Aggregate-Prone Regions in the Primary Amino Acid Sequence
The primary amino acid sequences of antibodies play a crucial role in determining their secondary, tertiary, and quaternary protein structures, which in turn influences their propensity to aggregate.
APRs can be located in the complementarity-determining regions (CDRs) of the Fab domain. These regions are hypervariable in their amino acid sequences and are the core component for antigen binding. CDRs contain aggregation-prone motifs rich in hydrophobic, aromatic, and uncharged amino acids, such as Gln, Asn, Tyr, Trp, and Phe. Stretches of hydrophobic residues including Val, Ile, and Leu have high aggregation propensity due to hydrophobic side chains. However, charged residues (Asp, Glu, Lys, His, and Arg) tend to not appear in APRs, consistent with the notion that charged residues aid in protein solubility.
In IgGs, more ARPs are found in the Fc than in the Fab region. Fc regions contain conserved stretches of hydrophobic residues in the lower hinge region, which leads to self-association and aggregation.
Aggregate-Prone Structural Motifs in Antibodies
Conformations and structural motifs can give rise to antibody aggregation through covalent and non-covalent interactions between exposed residues and side chains. One common structural motif found in antibodies, present in both Ig heavy and Ig lambda light chains, is the β-strand sandwich fold. The exposed edge of β-stands have been shown to trigger the formation of amyloid-like fibril aggregates, which has been observed in antibody therapeutics such as trastuzumab.
Additionally, structural characteristics such as non-native disulfide bonds can significantly alter and deteriorate antibody stability and increase aggregation potential. When cysteine residues remain unpaired, free sulfhydryl groups can lead to intramolecular scrambling and intermolecular crosslinking, thereby promoting antibody aggregation.
Post-Translational Modifications That Impact Aggregation Propensity
PTMs of antibodies are common to modulate their biological activity, but may also have significant impacts on antibody stability and aggregation propensity (Figure 2).
- Antibody Glycosylation: Glycosylation has been found to increase conformational stability and reduce aggregation. In IgGs, the highly conserved N-glycosylation on the CH2 domain of the Fc region reduces aggregation propensity and protects against protease degradation.
- Deamidation: Deamidation of asparagine in the Fc region is a common antibody modification and has been shown to increase antibody aggregation and reduce colloidal stability in a pH-dependent manner.
- Isomerization: Aspartate isomerization is strongly associated with structural instability of mAbs, leading to denaturation and protein aggregation.
- Oxidation: Methionine oxidation is primarily observed in the Fc region, and results in decreased antibody conformational stability and increased aggregation propensity, especially at higher temperatures. Tryptophan oxidation in the CDRs also increases the potential for protein aggregation.
It is important to emphasize that the impacts of chemical modifications and PTMs on aggregation and stability are specific to each antibody, as well as the solution and formulation conditions. To better understand the mechanisms that affect stability and solubility, further PTM characterization, stability, and aggregation analysis should be conducted on the antibody candidate.
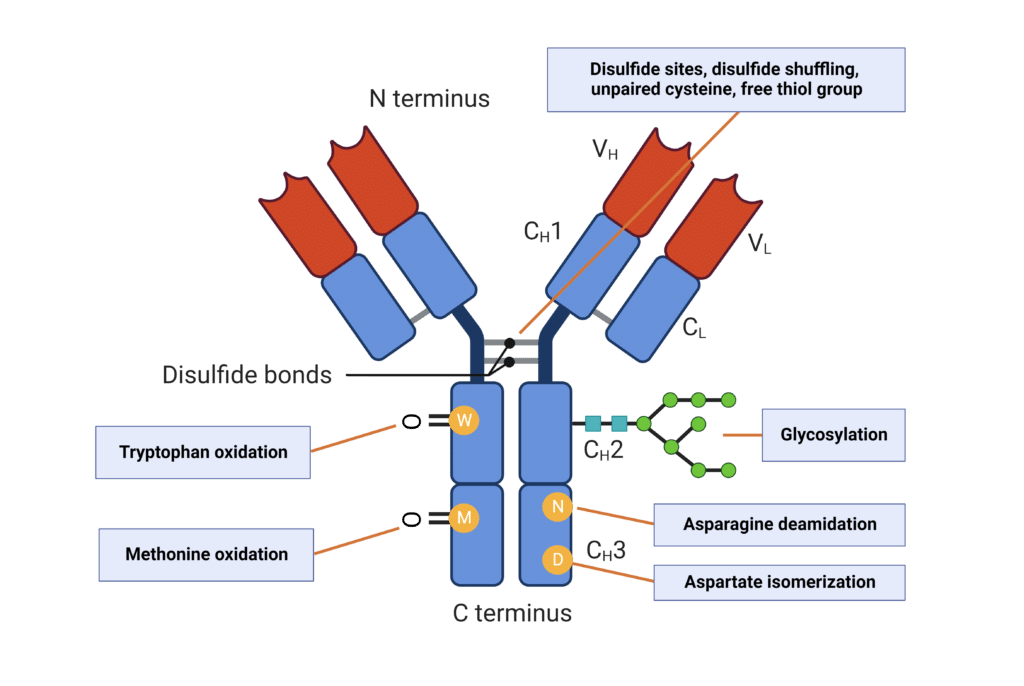
Figure 2. Post-translational modifications that influence antibody aggregation.
Engineered Antibody Formats That Influence Aggregation
With the rise of antibody engineering capabilities and the introduction of new antibody formats, certain formats have demonstrated an increased susceptibility to aggregation.
Antibody Drug Conjugates
Aggregation of antibody drug conjugates (ADCs) is not only related to the parent mAb, but it is also influenced by the conjugation chemistry and the type of linker-payload used. Conjugation reactions that link the payload to the mAb involve cysteine or lysine side chains, resulting in free sulfhydryl groups of cysteine residue and an exposed maleimido group from the linker. Intermolecular crosslinking between these groups can promote aggregation of ADCs. Additionally, the nature of the linker itself can be hydrophobic, providing an additional mechanism for ADC aggregation.
Single Chain Variable Fragments and Nanobodies
Single chain variable fragments (scFv) consist of the variable regions of the heavy and light chain fused into a single polypeptide chain. Hydrophobic residues previously buried in the variable and constant interface become exposed to the solvent, contributing to protein aggregation and instability. Sequence analysis of scFv has indicated that mutagenesis of hydrophobic residues to hydrophilic residues significantly improves in vivo folding and prevents aggregation.
Interestingly, nanobodies derived from camelid antibodies have been observed to be aggregation-resistant and are capable of reversible unfolding. This is due to a notable difference in hydrophilic residues at the interface where a variable light chain domain would typically be expected.
Strategies to Reduce Antibody Aggregation
Reducing the aggregation propensity of antibodies is critical in the development of therapeutic proteins to ensure their efficacy, and safety. Aggregation can lead to loss of biological activity and immunogenicity, potentially impacting patient health. Sequence and structural-based methods can be employed to mitigate and reduce aggregation propensity of antibodies.
Sequence-Based Approaches
Some sequence-based approaches to mitigate antibody aggregation include:
- In silico aggregation prediction algorithms can computationally predict ARPs, and consider hydrophobicity, charge distribution, and secondary structure propensities deduced from the primary amino acid sequence.
- Amino acid optimization can be employed to introduce hydrophilic residues and acidic residues within APR regions, as they are less likely to promote aggregation.
- CDR engineering by modifying amino acid sequences that flank the CDRs to reduce aggregation without impacting affinity and binding specificity.
Deriving the amino acid sequence of the antibody candidates is critical for effective sequence engineering, as well as being able to utilize in silico aggregation-prediction algorithms (Figure 3).
It’s important to remember that the same biophysical forces that influence antibody aggregation also dictate proper protein folding. A careful balance is required between optimizing antibody developability and maintaining antibody structure and function.
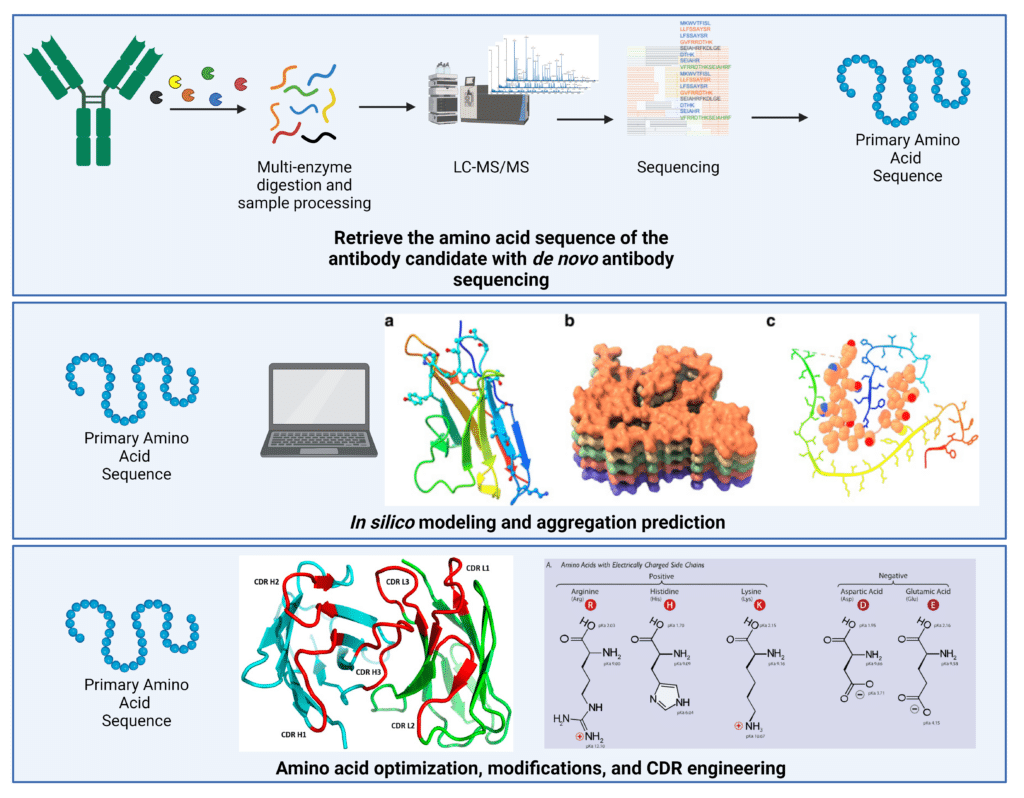
Figure 3. Sequence-based methods to reduce antibody aggregation. Since aggregation is primarily affected by the amino acid sequence, obtaining the antibody sequence is a critical first step. De novo antibody sequencing using liquid-chromatography with tandem mass spectrometry can derive the entire amino acid sequence with 100% coverage and accuracy. This primary amino acid sequence can then be utilized in in silico modeling and aggregation prediction, as well as rational amino acid optimization and CDR engineering to improve developability. In silico modeling figure adapted from: Prabakaran et al., 2021
Structural-Based Approaches
Structural-based approaches focus on the molecular interactions and conformational characteristics of the antibody that contribute to aggregation. Engineering modifications to the antibody structure can stabilize its conformation and limit the exposure of APRs in solution. Some structure-based methods for improving antibody aggregation are:
- Introducing stabilizing interactions within the antibody structure, such as adding disulphide bonds and modifying glycosylation patterns.
- Epitope mapping to identify antibody binding sites, and conformational changes to help identify potential APRs.
Biophysical Characterization of Proteins
Rapid Novor supports therapeutic antibody developability through de novo protein sequencing and antibody characterization services, such as glycan profiling and HDX-MS epitope mapping.
To learn more about our services and how we can accelerate your antibody development, contact our scientists.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

