Written by:
Written by:
Vanessa Yoon Calvelo, PhD
Published: July 11, 2022
What is Gene Therapy?
Gene therapy, in its simplest form, is the introduction of genetic material into target cells to treat or prevent clinical indications by correcting or supplementing the defective gene. Both ex vivo strategies – harvesting target cells for genetic modification followed by infusion back into the patients – and in vivo strategies – delivering genetic material directly into the target cells of patients – have been employed to achieve gene therapy (Figure 1).
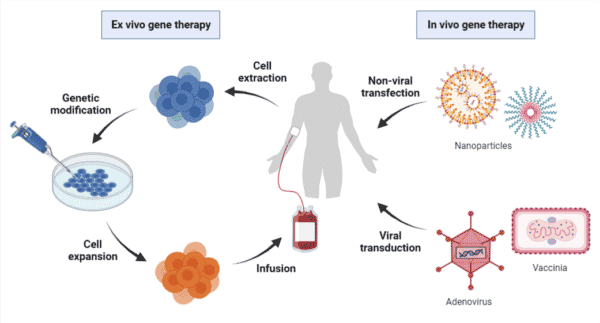
Figure 1. Overview of ex vivo and in vivo gene therapy processes. Ex vivo gene therapy requires the extraction of patient cells for genetic modification followed by infusion back into the patient. In vivo gene therapy involves the direct delivery of therapeutic genes via transfection of non-viral vectors or transduction of viral vectors.
The potential for gene therapy applications can include:
- The delivery of functional genes for clinical indications caused by gene defects, such as haemophilia, which results from mutations in clotting factor genes.
- The delivery of nucleic acids (ie. small interfering RNAs) knocking down overexpressed genes for clinical indications caused by the accumulation of toxic misfolded proteins, such as Huntington disease.
- The delivery of protein products that improve disease phenotypes for clinical indications that are caused by unknown gene mutations, such as antibodies for infectious diseases or calcium signaling proteins for heart failure.
Recent decades have seen therapeutic genes delivered to target cells in preclinical and clinical studies using viral vectors that have been engineered for gene delivery (Figure 1). It is important to note that non-viral vectors, such as nanoparticles, have also been employed for in vivo gene therapy; however, the scope of this article will be focusing on viral gene delivery platforms, in particular the adeno-associated viruses (AAVs).
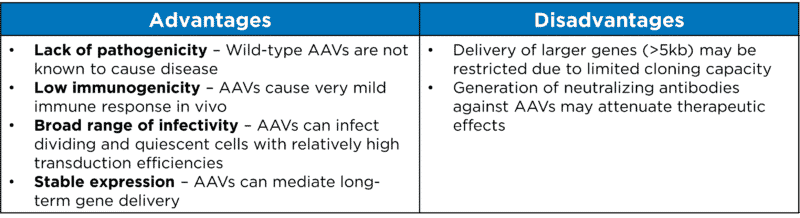
AAVs appear to be very attractive candidates for creating viral vectors for in vivo gene therapy. Some of the advantages and disadvantages of AAVs are listed in Table 1 below.
Table 1. Advantages and disadvantages of AAVs as viral vectors for gene delivery.
What are Adeno-Associated Viruses?
AAVs are small, non-enveloped viruses belonging to the genus Dependoparvovirus within the family Parvoviridae. AAV was first discovered in 1965 as a contaminant in laboratory preparations of adenovirus. It was named and classified accordingly since its life cycle depends on the presence of a helper virus, such as adenovirus, herpes virus, human papillomavirus, or vaccinia virus. To date, 108 isolates have been identified from multiple vertebrate species, including human and non-human primates, and classified into one of 13 different AAV serotypes (AAV1-AAV13). Wild-type AAVs naturally infect humans around 1-3 years of age; however, there are no known indications of AAVs associated with disease or illness.
AAV Composition and Structure
AAVs are composed of a protein capsid of ~26 nm in diameter with a triangulation number (T)=1 icosahedral symmetry (Figure 2). A total of 60 virion protein subunits (VPs) of 3 types – VP1, VP2, and VP3 in a ratio of 1:1:10, respectively – assemble the capsid by 2-, 3- and 5-fold symmetry-related interactions.
Packaged within the capsid is a single-stranded ~4.7-kb genome of either the sense or anti-sense strand (Figure 2). The genome comprises the rep and cap genes, which are flanked by two T-shaped inverted terminal repeats (ITRs). The single rep open reading frame (ORF) encompasses the viral origin of replication encoding the four Rep proteins – Rep78, Rep68, Rep52, and Rep40 – that are involved in AAV genome replication, packaging, and integration. The single cap ORF encodes the three capsid VPs, which are regulated and generated by transcription from the rare start codon ACG and alternative splicing. Located within the cap gene is the in-frameshifted aap ORF, encoding the assembly activating protein (AAP), which is essential for VP3s to assemble the capsid.
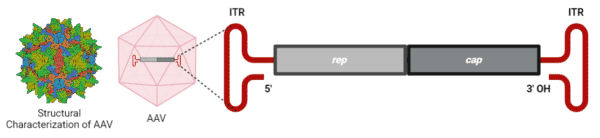
Figure 2. AAV2 capsid and genome. The AAV2 capsid is generated from 60 virion protein monomers symmetrically arranged in an icosahedral architecture. A ~4.7-kb genome is encapsulated within the capsid. Structural characterization of AAV2 was provided by RSCB PDB (#7RWT).
Engineering of AAVs for Gene Therapy
Engineering of AAVs have led to the production of recombinant AAVs (rAAVs), which are the leading platform for in vivo delivery of gene therapy. rAAVs consist of the same capsid sequence and structure as found in wild-type AAVs, but they differ in their genomes. In place of the AAV rep and cap genes, rAAV genomes have a therapeutic gene expression cassette, consisting of a promoter, a transgene, and a transcription termination signal, flanked by the two ITRs (Figure 3). The rep and cap genes are instead supplied by a trans-plasmid that is transfected into the host along with the rAAV genome and helper adenoviral genes. The complete removal of the viral coding sequences from rAAVs maximizes its packaging capacity (smaller genomes <5kb are effectively packaged more so than larger genomes >5 kb) and lowers their immunogenic and cytotoxic properties when delivered in vivo.
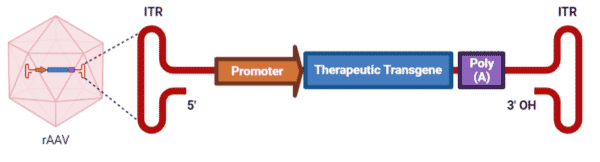
Figure 3. rAAV genome packaged within a wild-type AAV capsid.
Engineering AAVs for Improved Transduction
One of the determining factors for successful gene delivery is the transduction efficiency of rAAVs. The intracellular trafficking of rAAVs requires multiple cellular events that may abort at any given step. As a result, engineering strategies have been explored to optimize the capsid and rAAV genome for improvements in transduction efficiency (Figure 4).
Capsid Engineering
AAV capsids interact and bind to primary cell surface receptors and/or co-receptors, triggering the internalization step of the transduction pathway. Insights into the protein sequence, structure, and receptor-binding domain have enabled the rational design and enhancement of AAV capsids. A few common techniques for rational engineering include:
- Simple grafting of peptide sequences to bind to cell-type-specific receptors – for example, the introduction of integrin-binding peptide sequences onto the capsid surface, resulting in the re-targeting of AAV2 to resistant cells.
- Fusion of peptide sequences to confer enhanced binding capacities – for example, fusion of single-chain variable fragments (scFvs) onto the VP2 amino terminus to confer antibody-like recognition to cell surface markers.
- Site-directed mutagenesis of specific amino acid residues to disrupt capsid degradation – for example, mutagenizing surface-exposed tyrosines to increase transduction by inhibiting proteasomal degradation and facilitation of intracellular trafficking.
Other technologies employed in capsid engineering include directed evolution, a high-throughput, molecular method that mimics natural evolution through iterations of genetic diversification under artificial selection pressure; and in silico design, a computational method using high-throughput sequencing and bioinformatic tools to identify variable regions that tolerate manipulation.
rAAV Genome Engineering
Modifications to the ITRs, promoter, and transgene can all enhance rAAV transduction efficiency. Introducing mutations into the ITR sequences to generate specific self-complementary AAV replication intermediates can bypass the need for double-stranded DNA synthesis from the single-stranded rAAV genome, which is often the rate-limiting step in AAV transduction. Promoter-swapping from long and weak to small and strong cell type-specific promoters can increase the transcription of the transgene. Lastly, cell type-specific codon optimization of the transgene to augment mRNA production and translation can improve the transduction of larger transgenes.
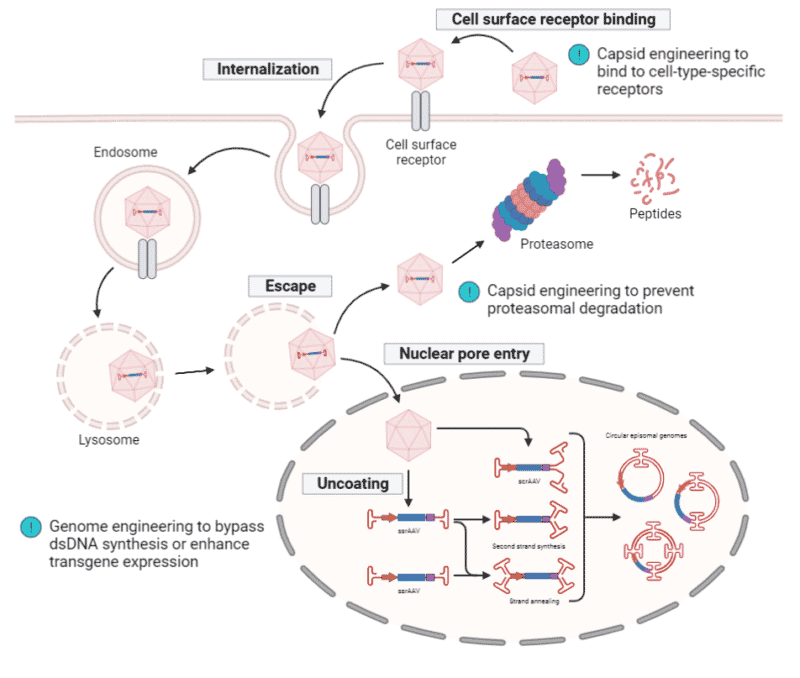
Figure 4. rAAV transduction pathway. rAAV binds to the cell surface receptor, triggering internalization and is trafficked through the cytosol within the endosome. Upon endosomal escape, rAAV can undergo proteasomal degradation or can be transported into the nucleus where uncoating of the capsid takes place. Either a single-stranded (ss) or self-complementary (sc) rAAV genome is released and transcription only begins upon the conversion of the ssrAAV genome into a double-stranded (ds) rAAV genome via second strand synthesis or strand annealing. The scrAAV genome immediately undergoes transcription since it is already in a double-stranded state. ITRs drive inter- and intra-molecular recombination to form circularized episomal genomes, allowing them to persist within the nucleus. Engineering of capsids have increased their binding specificity and affinity to cell surface receptors as well as disrupt proteasomal degradation. Engineering of genomes can bypass the need for dsDNA synthesis or enhance transgene transcription and translation.
Engineering AAVs for Improved Immunogenicity
Another determining factor for successful gene delivery is the immunogenicity of rAAVs. Approximately 40-70% of the human population have been exposed to wild-type AAVs and carry anti-AAV antibodies. Though rAAVs lack the viral genes and cannot synthesize viral proteins, they are assembled from wild-type capsid shells that can be recognized by the host immune response. It is, therefore, important to understand how antibodies interact with the AAV capsid to enable improvements in this gene delivery system (Figure 5).
Mapping Capsid Antigenic Epitopes
One approach to achieving a better understanding of the host antibody response towards AAVs is by mapping capsid antigenic epitopes. Each capsid VP subunit has nine variable regions on the surface that impact viral tropism and intracellular trafficking and are typically the domains recognized by neutralizing antibodies (NAbs). A common technology applied in the mapping of capsid epitopes is epitope searching, which focuses on the direct interactions between antibodies and capsid peptide sequences using three main strategies:
- Peptide scanning with short sequences that inhibit NAb binding – for example, scanning of the AAV2 capsid protein sequence using 91×15-mer peptides in an ELISA assay led to the identification of eight peptides that inhibit NAb binding
- Peptide insertion of short sequences generating AAV variants that evade NAb binding – for example, insertional mutations at amino acid positions 534 and 573 in the AAV2 capsid protein sequence demonstrated a reduction in NAb binding
- Site-directed mutagenesis of amino acids to identify specific residues involved in NAb binding – for example, screening of 57 alanine and 41 non-alanine substitutions led to the identification of six site-specific mutations that decreased neutralization.
Other technologies for epitope mapping of capsids have included directed evolution and structural biology. Directed evolution can involve error-prone mutagenesis to introduce random point mutations into the cap ORF, or DNA shuffling to randomly combine cap sequences from different AAV serotypes, leading to the generation of a genetically diverse library that is screened under the selective pressure of antibodies for neutralization-escaping variants. Structural biology often involves cryo-reconstruction of AAV capsids bound to fragment antigen-binding (Fab) regions that are generated from monoclonal antibodies to characterize the AAV antigenic structure.
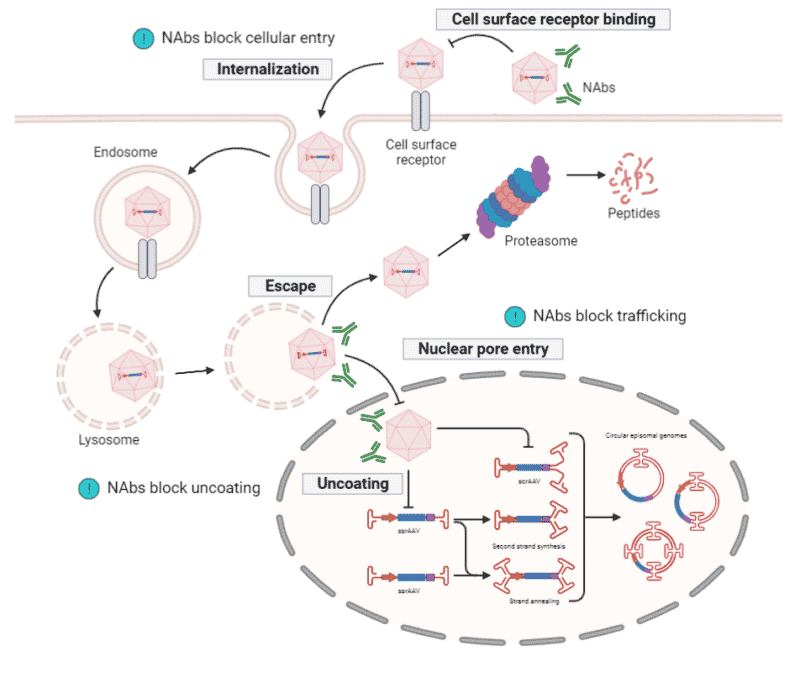
Figure 5. Antibody response towards rAAV transduction. Neutralizing antibodies (NAbs) bind to the capsid surface to prevent rAAVs from interacting with, and entering, target cells. NAbs can also block intracellular trafficking and capsid uncoating within the nucleus.
De Novo Protein Sequencing Applications in AAV Characterization and Development
The characterization and engineering methods discussed above have greatly progressed the AAV research field over the course of 20 years. As technologies and tools continue to improve and advance, novel ways to study these viral vectors for subsequent development into functional therapeutics seems to be within reach. One such technology is de novo protein sequencing, which is a mass spectrometry-based technique for the elucidation of amino acid sequences of proteins. Knowledge of protein sequences is one of the key factors in driving strategic engineering of AAVs.
Rapid Novor provides a suite of de novo protein sequencing services that can support the AAV characterization and development pipeline. Our REmAb® service can provide de novo sequencing of monoclonal antibodies, scFvs, and Fabs that are required for the strategic engineering of capsid proteins. Our REpAb® antibody discovery service can exploit the functional native immune response in patients as a means of identifying novel scFvs to equip AAVs with enhanced antibody-like recognition, or to develop anti-drug antibody assays.
Rounding out our protein sequencing services, we offer proteomic services for protein-protein interaction and kinetic analyses. HDX-MS service at Rapid Novor is a high-throughput, high-resolution method of choice for antigenic epitope mapping due to its capabilities in characterizing linear, conformational, and structural epitopes. Our new SPR service for antibody-antigen interaction and kinetic binding analysis can provide protein binding kinetic information for a complete understanding of the mechanisms behind AAV neutralization and immunity. To learn more about our services, reach out to our scientists for inquiries.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics