Written by: Genya Gorshtein, MSc
Written by: Genya Gorshtein, MSc
Published: July 17, 2025
Introduction
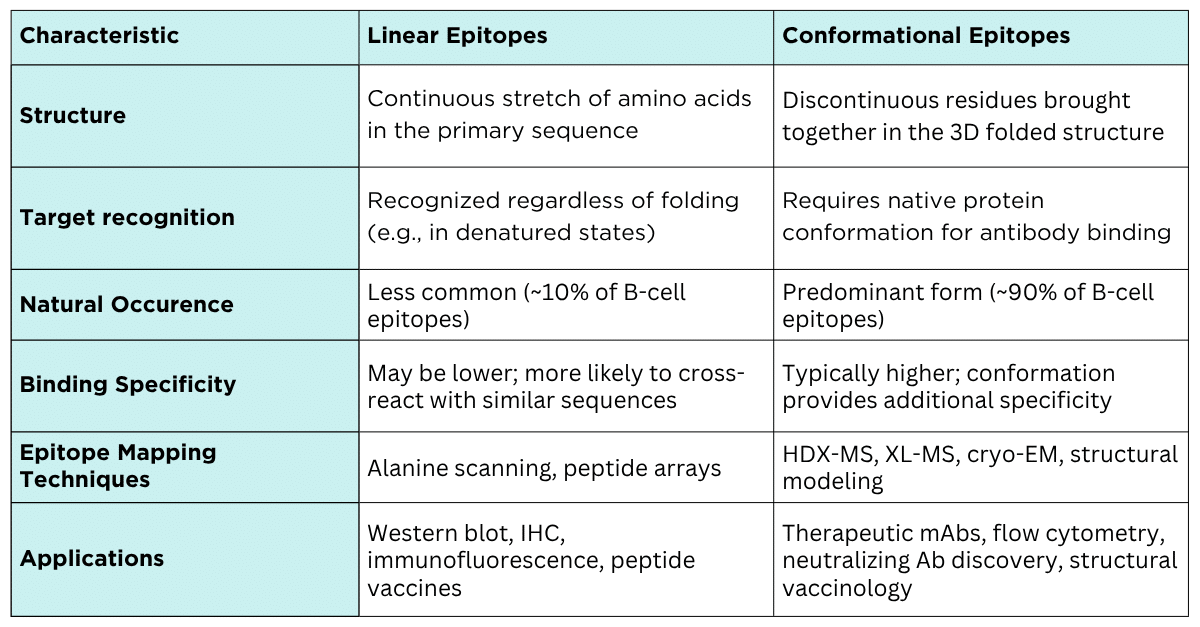
Antibodies recognize specific regions on antigens called epitopes. These can be broadly classified into linear (or continuous) and conformational (or discontinuous) epitopes.
While both types are important in immune recognition, the vast majority; up to 90% of B-cell epitopes are conformational, relying on the antigen’s precise three-dimensional shape rather than its linear amino acid sequence.
This structural complexity presents unique challenges for epitope mapping, requiring specialized techniques to accurately characterize the interaction between an antibody and its target.
Table 1. Summary of the characteristics of linear vs. conformational epitopes.
What is a Linear Epitope?
Linear epitopes are composed of a continuous sequence of amino acid residues in the antigen’s primary sequence (Figure 1). They remain recognizable by antibodies even when the protein is denatured, making them easier to study using methods that don’t rely on the protein’s native conformation.
These epitopes are often preferred in applications where the antigen is fully or partially denatured, such as Western blot (WB), immunohistochemistry (IHC), or immunofluorescence microscopy.
Advantages of linear epitopes include:
- Easier to map using techniques like alanine scanning mutagenesis.
- Do not require the antigen to retain its native 3D structure.
- Cost-effective and scalable for epitope mapping; synthetic peptides representing short linear sequences are sufficient.
Limitations include:
- May have reduced biological relevance in native or in vivo settings.
- Antibodies raised against linear peptides may fail to recognize the native form of the protein, particularly if the epitope is buried.
- Represent a minority of naturally occurring epitopes.
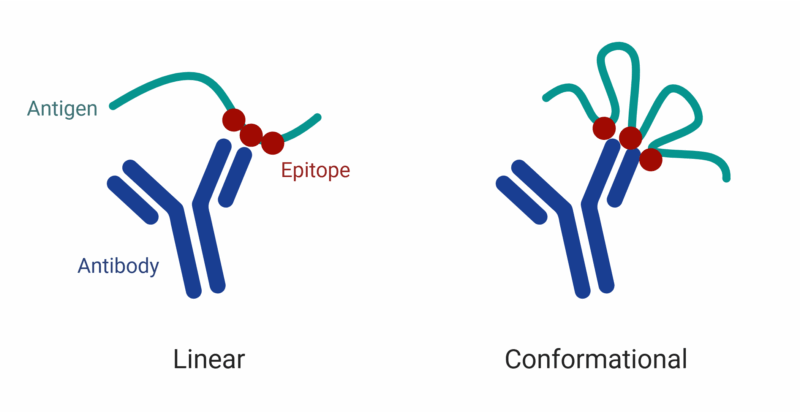
Figure 1. Linear vs. conformational epitopes. Linear epitopes consist of a continuous sequence of amino acids recognized by the antibody, while conformational epitopes are formed by discontinuous residues brought together by the folded 3D structure of the antigen.
Linear Epitope Mapping Techniques
Alanine Scanning Mutagenesis
Alanine scanning involves systematically substituting each amino acid residue within a suspected epitope region with alanine and evaluating the impact on antibody binding. Because alanine is small and uncharged, this technique helps identify critical side chains responsible for antigen-antibody interactions without significantly altering the backbone conformation.
- Strengths: Identifies individual residues essential for binding, relatively inexpensive and accessible technology; works best with preliminary structural data or binding predictions.
- Limitations: Best applied to known or suspected epitope regions; can be labour intensive; not suitable for conformational epitopes.
Peptide Arrays
Peptide arrays consist of overlapping synthetic peptides representing the linear sequence of the antigen, immobilized on a solid surface. These arrays are incubated with the antibody of interest to identify the binding regions.
- Strengths: Rapid and high-throughput; does not require the native protein.
- Limitations: May miss discontinuous or conformational epitopes; sensitive to peptide length and orientation.
Together, alanine scanning and peptide arrays provide accessible tools to map linear epitopes at both the residue and sequence levels. While they may not capture the complexity of conformational binding sites, these methods are effective when the target epitope is accessible and sequence-defined.
What is a Conformational Epitope?
Conformational epitopes consist of amino acids that are non-contiguous in the primary sequence but come together in the protein’s folded three-dimensional structure (Figure 1). These epitopes are typically lost when the protein is denatured, making their identification and characterization dependent on preserving the native conformation of the antigen.
Conformational epitopes are preferred in applications where the target protein remains in its native state, such as therapeutic antibody development, in vitro diagnostics and other lateral flow assays, flow cytometry, or in vivo functional studies.
Key applications include:
- Therapeutic antibody development, particularly for complex targets like membrane proteins.
- Neutralizing antibody discovery in infectious disease and cancer immunotherapy.
- Structural vaccinology, including epitope scaffolding and rational immunogen design.
Advantages of conformational epitopes include:
- Reflect the native, biologically relevant binding interaction.
- Typically associated with higher target specificity.
- Capture subtle conformational changes essential to function (e.g., receptor binding, activation states).
Limitations include:
- Cannot be mapped using denatured proteins or linear peptides.
- Require advanced techniques like HDX-MS, XL-MS, or cryo-EM for accurate characterization.
- Sensitive to changes in protein folding, including formulation conditions, pH shifts, and post-translational modifications.
Conformational Epitope Mapping Techniques
Hydrogen-Deuterium Exchange Mass Spectrometry (HDX-MS)
HDX-MS measures the exchange of hydrogen atoms with deuterium in backbone amides, which is protected by antibody binding. This technique reveals epitope regions by identifying areas of the antigen that become less solvent-accessible upon complex formation.
- Strengths: Native-state mapping; sensitive to subtle conformational changes, can reveal dynamic information.
- Limitations: Moderate resolution; requires careful experimental design and optimization.
Cryo-Electron Microscopy (cryo-EM)
Cryo-EM enables direct visualization of antibody-antigen complexes at near-atomic resolution. It is especially powerful for large, flexible, or membrane-bound proteins where crystallization is challenging.
- Strengths: High-resolution; ideal for complex targets.
- Limitations: Technically and computationally demanding; time and resource-intensive.
Crosslinking Mass Spectrometry (XL-MS)
XL-MS captures spatial proximity between antibody and antigen residues using chemical crosslinkers, followed by mass spectrometry analysis.
- Strengths: Maps epitopes in native-like states.
- Limitations: Lower spatial resolution; provides distance constraints rather than exact binding sites; potential for crosslinking-induced conformational artifacts.
Computational Structural Prediction
Computational methods predict 3D structures of antibodies and antigens to identify likely conformational epitopes. Techniques include homology modeling, molecular docking, and machine learning-based epitope prediction.
Strengths: Rapid, cost-effective, guides experimental design.
Limitations: Dependent on model accuracy; should be validated experimentally.
For a more comprehensive comparison of epitope mapping techniques, visit our article: Comparing Epitope Mapping Methods.
Antigen Design for Antibody Discovery: Linear vs. Conformational Epitope Considerations
Antigen selection plays a critical role in shaping the type and specificity of antibodies generated during antibody discovery. Whether the goal is to target linear or conformational epitopes, the structure and presentation of the antigen directly influence the outcome.
Full-Length Protein Antigens for Conformational Epitope Discovery
Full-length proteins are commonly used when the goal is to generate antibodies against conformational epitopes. These proteins preserve the native three-dimensional structure of the antigen, allowing for the recognition of complex, folded regions.
However, one drawback of using full-length proteins is the potential for cross-reactivity. Antibodies may bind conserved regions shared with other proteins, reducing specificity and increasing the risk of off-target effects. To address this, functional enrichment and depletion strategies, such as depleting antibodies that bind to a homologous protein or sequence, can help filter out non-specific binders. When combined with polyclonal antibody sequencing, these approaches enable the identification of antigen-specific antibodies from enriched populations targeting defined regions, enhancing both precision and downstream utility.
Short Peptide Antigens for Linear Epitope Discovery
Short linear peptides of 10-20 amino acids, coupled to a carrier protein can be used to generate antibodies that target unique, sequence-specific regions of the protein. By selecting peptides from regions with minimal sequence homology to other proteins, researchers can create highly specific antibodies. However, anti-peptide antibodies often struggle to recognize the native protein, as the unstructured nature of short peptides may not accurately mimic the conformational context of the full antigen.
Protein Fragments as a Balanced Antigen Design Strategy for Discovery
An effective middle ground is the use of protein fragments, typically 50 to 150 amino acids in length, that span unique regions of the target protein. These fragments are large enough to retain some secondary and tertiary structure, increasing the likelihood that antibodies raised against them will recognize the native protein. At the same time, selecting a fragment from a region with low sequence similarity to other proteins helps reduce cross-reactivity. This approach offers a practical balance between specificity and structural relevance, making it a valuable strategy in antibody discovery.
However, a potential limitation is the cost associated with synthesizing these larger fragments, which can be significantly higher than ordering short peptides or purchasing commercially available full-length proteins, especially when post-translational modifications or expression in specific systems are required.
Protein Interaction Analysis Service with HDX-MS
HDX-MS offers a label-free approach for mapping protein-protein interactions at high resolution. Our HDX-MS service enables both epitope and paratope mapping, revealing interaction interfaces, conformational changes, and binding sites with localized precision. Binding interactions are captured in near-native conditions, preserving secondary and tertiary structure, ideal for mapping both conformational and linear epitopes.
Our team includes PhD experts with over 7 years of experience in HDX-MS, working with a variety of analytes.
Contact our scientists to learn more about epitope mapping using HDX-MS.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics