Introduction
Enzyme-linked immunosorbent assays (ELISA) are widely used in therapeutic antibody development, from early screening to immunogenicity testing. However, ELISAs often underestimate binding affinity, especially for antibodies with slow association or fast dissociation rates.
Without kinetics data, it’s impossible to define the incubation time needed for ELISA to capture true affinity. Surface plasmon resonance (SPR) fills this gap by measuring binding in real time, yielding: association and dissociation rates, the equilibrium constant (KD), and the time to equilibrium (tequil). These parameters are essential for establishing an appropriate ELISA protocol with sufficient incubation time to ensure accurate and reproducible affinity measurements.
This case study shows how SPR-derived kinetics can guide ELISA optimization, helping ensure accurate affinity measurements and fit-for-purpose assay conditions.
Comparing ELISA and SPR for ADA Detection
A 2021 study by Beeg et al. compared ELISA and SPR results in 76 patients receiving infliximab for inflammatory bowel disease. While both methods produced similar serum drug concentrations, they differed significantly in detecting anti-drug antibodies (ADA).
All 14 samples identified as ADA-positive by ELISA were also positive by SPR; however, the absolute ADA levels measured by SPR were 7 to 490 times higher, with no correlation between the two methods. Furthermore, SPR detected ADA in 8 additional patients who were considered ADA-negative by ELISA.
Why the big difference?
The issue comes down to binding strength (affinity, KD) and how long the antibodies stay attached (dissociation rate, kd). ELISA uses a high-affinity commercial antibody with a very low dissociation rate (kd) as a reference or calibrator, and remains bound throughout the assay, even during long incubation and wash steps. This gives a stable, strong signal.
In contrast, the patient’s ADA usually binds more weakly and dissociates faster. As a result, much of the patient’s ADA is no longer bound when detection occurs, resulting in a reduced or missing signal and an underestimation of ADA levels.
SPR overcomes this limitation by measuring binding in real time, capturing both fast and close disassociation rates, providing a more accurate and comprehensive measurement of antibody binding.
Why Equilibrium is Key to Reliable Affinity and KD Values
A 2020 analysis by Jarmoskaite et al. at Stanford reviewed 100 studies reporting binding affinities. The study revealed that 70% of the papers failed to confirm equilibrium; a critical requirement for accurately determining KD values.
Despite the biological likelihood of long equilibration times, nearly 90% of the reviewed studies used incubation times of one hour or less. Yet, studies on protein complexes have shown that full equilibration can take many hours.
To reliably measure KD, it’s essential that the system reaches equilibrium at which the rate of binding equals the rate of dissociation, and the amount of bound complex remains stable over time. If measurements are taken too soon before equilibrium, the results underestimate how much binding would actually occur, leading to artificially high KD values and likely underestimate the true strength of interaction.
Case Study: Comparative Analysis of Affinity by ELISA vs SPR
In a prior alpaca antibody discovery study, several high-affinity clones were sequenced and identified against a monoclonal antibody antigen. Clones R4 and R9 were selected for detailed binding analysis by both ELISA and SPR.
Candidates R4 and R9 were first analyzed by ELISA (Figure 1), followed by kinetic analysis by SPR (Figure 2).
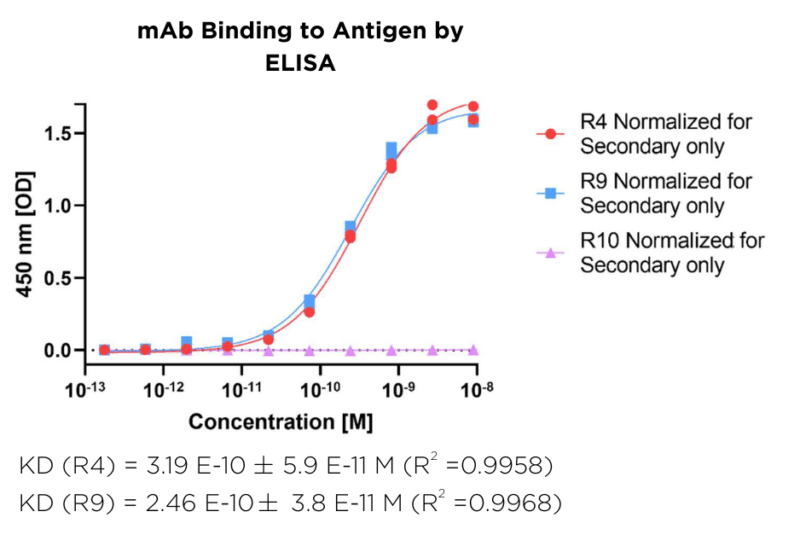
Figure 1. ELISA-based binding analysis of R4 and R9. Binding curves were fitted to determine apparent affinity, with goodness of fit assessed by the correlation coefficient (R² > 0.95).
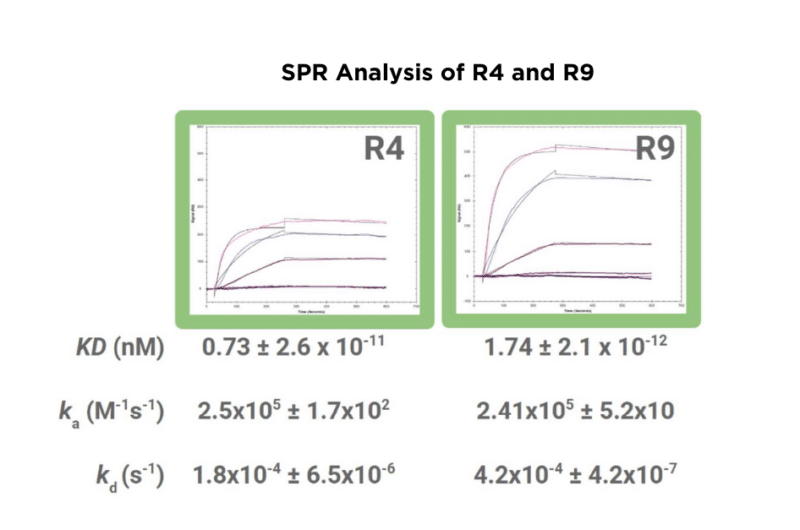
Figure 2. SPR analysis of R4 and R9 binding to the monoclonal antibody antigen. Sensorgrams and corresponding kinetic parameters (KD, ka, and kd) are shown, highlighting real-time binding interactions and affinity measurements.
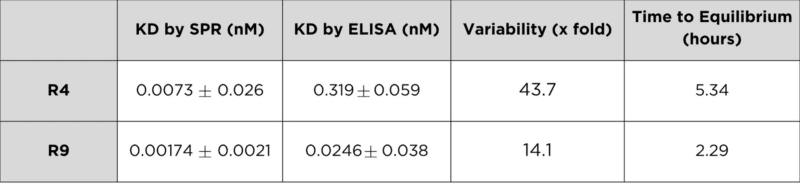
ELISA reported KD values that were 43.7-fold higher for R4 and 14.1-fold higher for R9 compared to SPR, significantly underestimating binding affinity (Table 1). This discrepancy suggests that equilibrium binding was not achieved during the ELISA incubation.
As an endpoint assay, ELISAs do not capture the dynamic nature of molecular interactions or confirm whether equilibrium has been reached. In contrast, SPR measures binding in real time, providing a complete kinetic profile.
Based on SPR kinetic analysis, the time to equilibrium (tequil) was calculated to be 5.34 hours for R4 and 2.29 hours for R9. If, for example, the ELISA for R4 were incubated for at least 5.34 hours, the measured KD would likely more accurately reflect the true affinity observed by SPR.
Table 1. Summary of KD values obtained through SPR vs. ELISA.
Optimizing ELISAs with SPR
Accurate affinity measurements by ELISA depend on reaching binding equilibrium, defined as tequil, the minimum incubation time required for a system to equilibrate. Without meeting this condition, the resulting affinity from ELISA is unreliable.
However, tequil cannot be predicted; it must be determined through kinetic analysis using SPR or a similar orthogonal method. SPR provides the association and dissociation rate constants that define equilibrium binding, making it the only reliable approach for calculating the true affinity constant (KD).
Once the kinetic profile is known, ELISA protocols can be rationally designed using the appropriate incubation time, ranging from minutes to even days, ensuring equilibrium is reached. This benchmark allows ELISA to be effectively used for relative comparisons, such as screening affinity-improved mutants or assessing batch-to-batch variation. However, ELISA should never be relied upon as the sole or primary method for measuring affinity.
In this way, SPR not only provides accurate kinetic data but also serves as a powerful tool to improve the performance and interpretability of ELISA-based assays.
SPR Analysis with Rapid Novor
Rapid Novor’s Surface Plasmon Resonance service delivers detailed kinetic profiling that can complement traditional assays like ELISA.
Whether you’re validating lead candidates or troubleshooting binding discrepancies, our platform provides real-time, quantitative data, offering both rapid SPR screening or full kinetic characterization performed in triplicate for robust, reproducible results.
Contact our scientists to learn more.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics