Written by:
Written by:
Genya Gorshtein, MSc
Published: November 25, 2022
Introduction
Small-molecule drug development is aimed at inhibiting disease-promoting protein function through occupancy-driven protein inhibition. A major caveat of this therapeutic development is the limitation of ‘druggable’ targets, being proteins with enzymatic or receptor function where an active or regulatory site can be occupied.
An emerging class of therapeutics called proteolysis-targeting chimeras (PROTACs) promotes degradation of the protein of interest (POI) by manipulating endogenous cellular proteostasis. This heterobifunctional molecule bridges the POI with an E3 ligase, resulting in its subsequent polyubiquitination and proteasome-dependent degradation.
This class of drug is particularly interesting, as the E3 ligase recruiting unit is conjugated to a protein-binding moiety that binds to the POI, such as a monoclonal antibody (mAb). Here, we discuss the unique and favourable properties of mAb conjugated PROTACs (abPROTACs) as novel pharmacological agents.
General PROTAC Structure and Function
The heterobifunctional PROTAC binds to the POI at one end while the other end binds to an E3 ligase to form a ternary complex (Figure 1). E3 enzymes catalyze a polyubiquitin signal on the POI, which signals it for degradation by the 26S proteasome. While the POI is degraded, the PROTAC molecule can be recycled for additional rounds of targeted degradation.
The PROTAC molecule consists of three components:
- Protein binding moiety – can be small molecule, peptide, or mAb based. Designing the targeting peptide requires epitope mapping through HDX-MS to identify key protein-protein interactions within the POI-peptide complex. Phage display technology, hybridoma, and B cell sequencing are common techniques for antibody generation. At Rapid Novor, we have introduced a fourth modality for antibody generation and discovery: polyclonal antibody sequencing.
- E3 ligase recruiting ligand – redirects protein degradation by recruiting E3 ligase. E3 ligases catalyze the direct transfer of a ubiquitin residue to the E3-bound substrate, the POI. While there are over 600 E3 ligases in the human genome, Von Hippel-Lindau (VHL) or Cereblon (CRBN) are the most common ligands used for PROTAC development due to their specificity and high-affinity targets.
- Linker – connects the protein binding moiety to the E3 recruiting ligand. Linker length, chemical composition, and attachment point affects the efficacy and the affinity of the PROTAC to its target. Glycine, serine, and polyethylene glycol (PEG) are commonly incorporated into linkers, taking advantage of affinity, spatial considerations, and hydrophilicity.
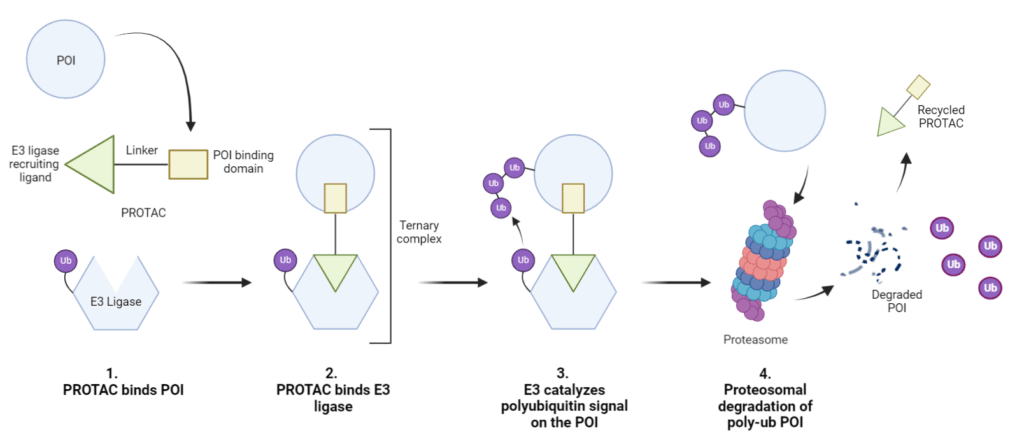
Figure 1. Structure and function of the basic PROTAC molecule. The PROTAC molecule binds the POI via the protein binding moiety and recruits a E3 ligase via the E3 ligase recruiting ligand. Following ternary complex formation E3 ligase catalyzes the addition of a polyubiquitin signal on the POI, signaling it for degradation by the proteasome.
Antibody-Conjugated PROTACs
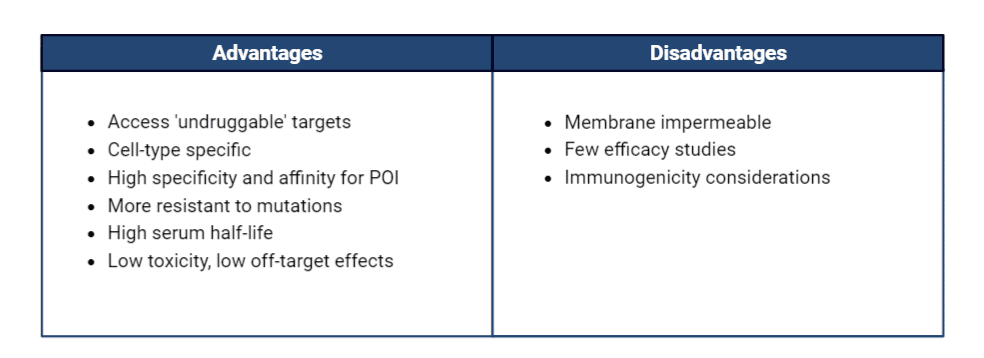
Traditional PROTACs utilize small molecules as the protein-targeting moiety, which are dependent on the availability of binding pockets within the POI. The benefit of peptide and mAb based PROTACs is the ability to engineer binders with high affinity to POI epitopes, while avoiding the limitation of shallow binding pockets on the POI. AbPROTACs pose several advantages over previous PROTAC technologies, described in Table 1.
Table 1. Advantages and disadvantages of abPROTACs over small-molecule PROTACs.
Small molecule and peptide based PROTACs are unable to discriminate between different cell types, and as such may contribute to off-target effects in healthy tissues. A unique characteristic of abPROTACs is their ability to promote selective protein degradation in a specific cell type through cell surface antigen recognition. The mechanism of action of an abPROTACs delivery is similar to that of an antibody-drug conjugate (ADC), such that binding of the antibody moiety to its cell-surface antigen results in endosomal internalization and subsequent drug delivery (Figure 2, left). AbPROTACs can be considered as an alternative approach to ADCs, as their catalytic activity may circumvent ADC-related dose-limiting toxicities due to potent cytotoxic payloads.
Expanding AbPROTACs Through Antibody Engineering
Expanding therapeutic targets of AbPROTACs can be achieved through protein engineering strategies. Development of bispecific abPROTACs can target membrane bound POIs for degradation by bridging it with a membrane bound E3 ligase (Figure 2, right). For example, a bispecific abPROTACs targeted programmed death ligand 1 (PD-L1) for lysosomal degradation by recruiting and binding membrane E3 ligase RNF43. AbPROTACs can be further optimized by exploring different IgG formats that range in flexibility, orientations, and fragment engineering such as scFv.
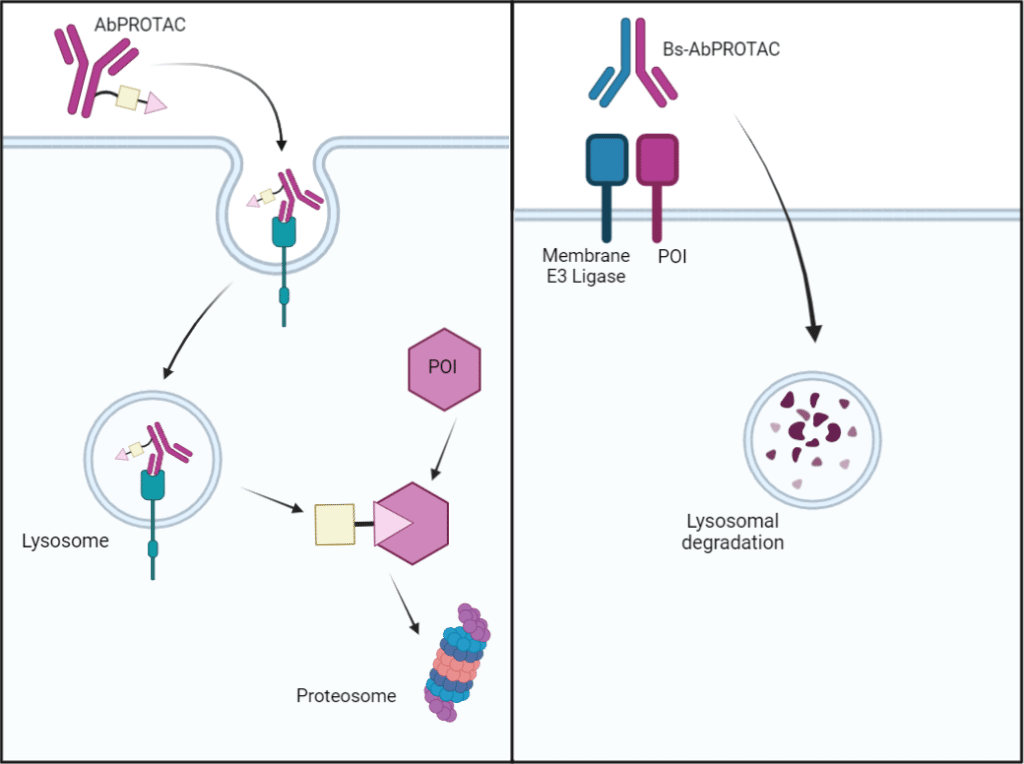
Figure 2. Methods of action of abPROTACs. On the left, the mAb-conjugated PROTAC recognizes and binds to its antigen present on the cell surface. This complex is internalized via endocytosis, releasing the PROTAC molecule into the cytoplasm. The PROTAC binds to the POI and E3 ligase, promoting targeted degradation of the POI. On the right, a bispecific PROTAC binds and bridges the cell surface POI with a membrane-bound E3 ligase. Following internalization via endocytosis, the POI is targeted for degradation via the membrane-bound E3-ligase.
Developing AbPROTACs with De Novo Antibody Sequencing and Proteomics
Antibodies have become increasingly common in the therapeutic landscape against cancer and various infectious diseases. The ability to exploit the unique characteristics of antibodies in facilitating the delivery of pharmacological agents, such as PROTACs, to target cells has significant potential for its application to novel targets. At Rapid Novor, we support antibody drug discovery through de novo antibody sequencing and proteomics.
De novo protein sequencing of monoclonal and polyclonal antibodies allows for the evaluation of lead candidates with key insights into relevant CDR and framework sequences for antibody engineering. Rapid Novor utilizes a well-established mass spectrometry-based antibody sequencing service to sequence antibodies directly from the mass spectral data, without reference to genomic sequence information.
Our proteomic services for analyzing protein-protein interactions augment our protein sequencing services for abPROTACs design. HDX-MS epitope mapping service can characterize antibody-antigen protein complexes, to accurately determine linear, conformational, and structural epitopes. SPR kinetic binding analysis provides insights into the binding affinity, specificity, and association and dissociation rates of the abPROTACs to its target antigen. Kinetic analysis is critical during PROTAC design as ligands, linkers, and protein binding moieties can impact binding interactions and ternary complex formation.
To learn more about our services, reach out to our scientists.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics