 Written by Genya Gorshtein, MSc
Written by Genya Gorshtein, MSc
Updated: January 27, 2023
(Published: August 31, 2022)
How is Antibody Diversity Generated?
Without the presence of an antigen, the human immune system can generate more than 1012 different antibodies and as many as 1018 in response to a foreign antigen. The mammalian immune system has evolved to generate more antibodies than it has genes for through mechanisms such as gene segment joining and somatic hypermutation (Figure 1). These mechanisms drive antibody diversity allowing the immune system to quickly detect and neutralize pathogens and fine tune the immune response.
Which Steps Contribute to the Generation of Antibody Diversity?
Combinatorial joining of gene segments, junctional diversification, and heavy and light chain pairing gives rise to antibody diversity during B cell development in the bone marrow, prior to interactions with antigens (Figure 1). Somatic hypermutation and class switch recombination generates antibody diversity in activated B cells after antigen stimulation. B cells are preferentially selected based on their affinity for the target antigen, thereby promoting affinity maturation.
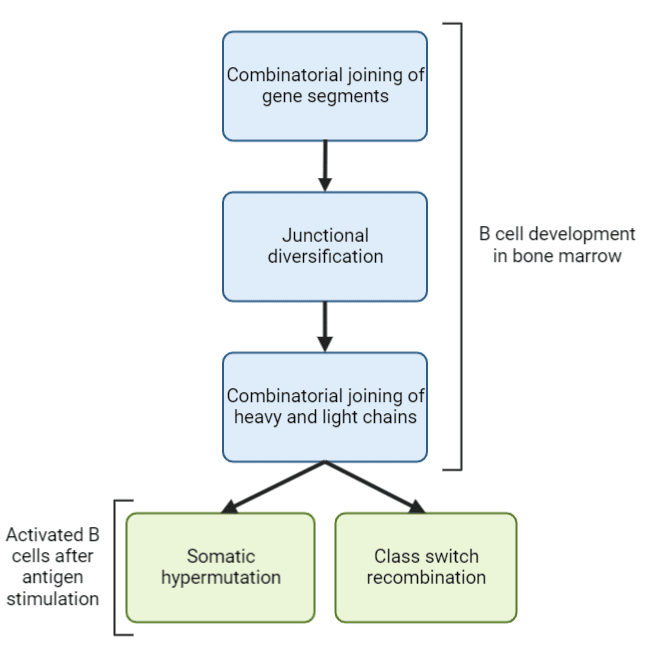
Figure 1. Main mechanisms of generating antibody diversity.
Antibody Loci and V(D)J Recombination
The antibody loci contain various gene segments that rearrange in developing B cells to generate a highly variable repertoire of antibodies. The variable region of the light-chain (LC) is assembled from two gene segments – a long variable (V) segment and a short joining (J) segment (Figure 2). The heavy chain (HC) variable region is assembled from a V segment, J segment, and a diversity (D) segment.
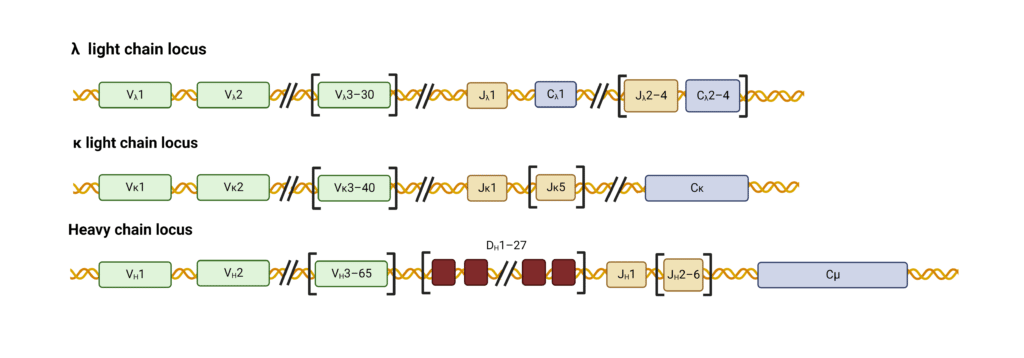
Figure 2. Organization of gene segments in human heavy and light chain loci.
Recombination and joining of V, D, and J segments create a functional variable immunoglobulin region (Figure 3). For human κ LCs, 40 V segments can combine with any of the 5 J segments resulting in 200 possible combinations encoded by this pool. The genes for human λ LCs contain 30 V segments and 4 J segments resulting in 120 possible variable λ regions. HCs contain 65 V segments that can join with any of the 6 J and 27 D segments to form roughly 11,000 possible variable HC regions.
A process called site-specific recombination mediates V(D)J recombination. Chromosomal DNA double stranded breaks are introduced by RAG recombinase. This enzyme complex binds and cleaves the DNA at conserved sequences that flank each V, D, and J segment. Selected V, D, and J segments are rearranged and joined to form the V(D)J exon. DNA ends are repaired by DNA repair enzymes, resulting in deletions or inversions of gene segments.
Nucleotides are often lost or inserted at the joining sites during recombination resulting in a frameshift mutation. This process known as junctional diversification introduces an additional level of antibody diversity in the variable region, specifically in the third complementarity-determining region (CDR3). The formed V(D)J exon is transcribed and translated into a functional HC or LC (Figure 3).
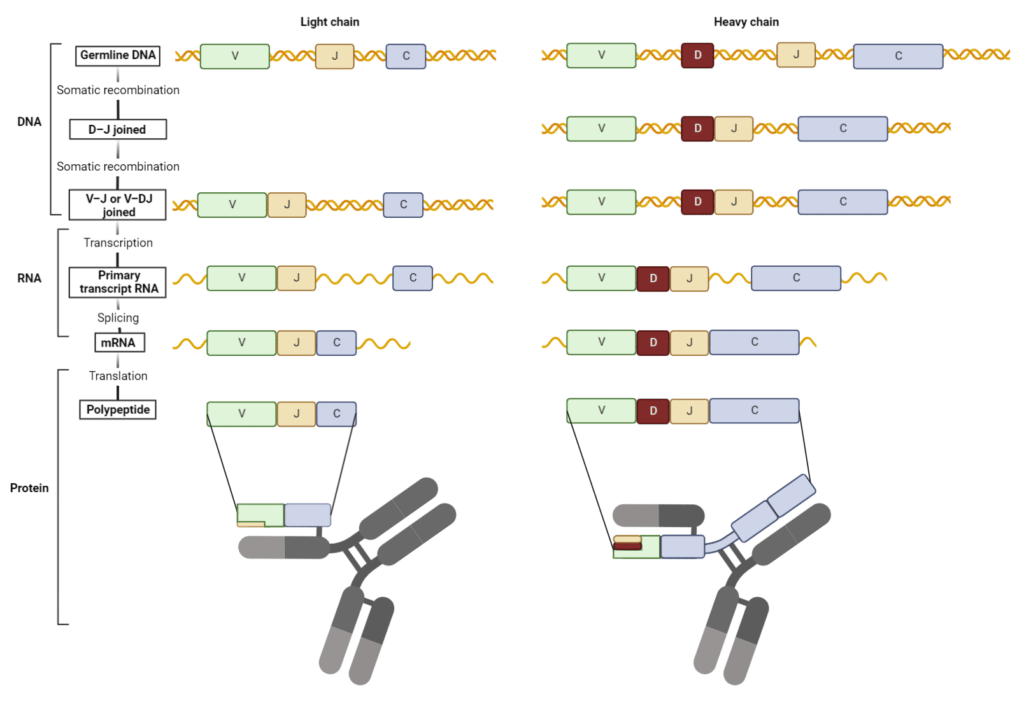
Figure 3. V(D)J recombination forms the variable region of heavy and light chains. (Adapted from Janeway et al. 2001)
In a single B cell, any possible HC can be recombined and produced together with any possible LC. Since both H+L chains provide antigen specificity at the antigen-binding site, this results in over 3 million unique antibodies. Combinatorial diversity of the antigen-binding site is achieved through different combinations of segment joining and H+LC pairing.
Somatic Hypermutation
Somatic hypermutation (SHM)provides an additional level of antibody diversity after V(D)J recombination. Point mutations by somatic hypermutation increase variation into the variable region and occur a million-fold more frequently than other genetic mutations. Double stranded DNA breaks are introduced into variable regions, eliciting DNA damage response pathways that facilitate error-prone repair and resulting in mutant antibodies.
SHM facilitates the progressive increase in antibody affinity against the antigen known as affinity maturation (Figure 4). Some immunoglobulin mutants bind antigens better than others, while some mutants produce non-productive rearrangements. Mutations in the framework regions of the variable domain tend to be selected against as they don’t enhance antigen-binding and alter the basic antibody structure. Mutations that enhance antigen-binding tend to be clustered in the CDR regions. Hypermutated B cells undergo a selective process in germinal centers where B cells compete for various signals in an affinity-dependent manner, thereby outcompeting lower-affinity B cell clones. Strong-affinity binders are selected for and proliferate and mature into antibody-secreting cells, whereas lower affinity clones are eliminated by apoptosis.
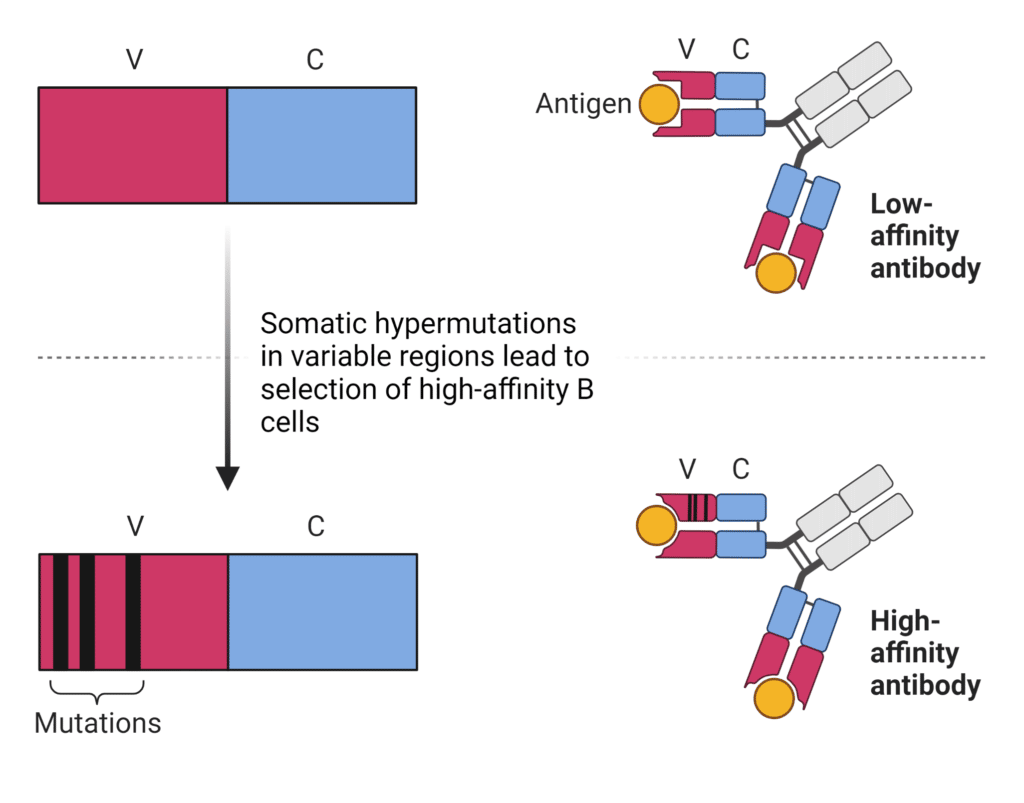
Figure 4. Somatic hypermutations result in higher-affinity antibodies.
Class Switch Recombination
During B cell development, cells switch between generating one class of antibody to another, a process known as class switch recombination (CSR). IgM is the first immunoglobulin to be synthesized by B cells, which are inserted into the plasma membrane as a B cell receptor (BCR). When B cells leave the bone marrow, they begin synthesizing IgM and IgD molecules embedded within the plasma membrane. Once these B cells interact and bind the antigen through the BCR, this results in a switch from membrane bound IgM to soluble IgM. As the immune response progesses, several class switching events occur to generate IgG, IgE, or IgA antibodies generating a secondary antibody response through memory B cells.
The constant region of the HC determines the antibody class. As such, CSR occurs through intrachromosomal deletions of the switch (S) region within the HC constant region. CSR is different from V(D)J recombination in that CSR occurs after antigen stimulation, is activated by helper T cells and involves different enzymes that recognize different flanking sequences on the DNA. CSR only alters the constant region without influencing the antigen-binding site. As such, CSR distributes antigen-binding sites across various antibody classes and provides diversity in effector responses and biological functions.
Antibody Sequencing Service
The complex mechanisms of antibody diversification make it difficult to characterize the antibody repertoire at the genomic level. At Rapid Novor, we can sequence monoclonal and polyclonal antibodies using a proteomics-only approach with our monoclonal antibody sequencing service and polyclonal antibody sequencing service. By using our de novo protein sequencing software, we can define the antibody repertoires and monitor the real-time status of the immune response.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

