Abstract
The identification of disease-causing mutations has progressed our understanding of disease pathogenesis and progression. Insight is further provided from individuals that carry these mutations but otherwise remain asymptomatic or show a delayed onset of clinical indications. This was the case of one individual who was identified to carry the presenilin 1 (PSEN1) E280A mutation, along with two copies of the APOE3 Christchurch (APOE3ch) R136S mutation, from the world’s largest autosomal dominant Alzheimer’s disease (ADAD) kindred consisting of 1200 PSEN1 mutation carriers.
A team of researchers led by Yakeel Quiroz (Massachusetts General Hospital, Harvard Medical School), Eric Reiman (The Banner Alzheimer’s Institute, University of Arizona), and Joseph Arboleda-Velasquez (Schepens Eye Research Institute of Mass Eye and Ear, Harvard Medical School) identified one PSEN1 mutation carrier who did not develop mild cognitive impairment (MCI) until her seventies – nearly three decades after the typical age of onset – indicating relative resistance to Alzheimer’s disease dementia. The PSEN1 mutation phenotype is often displayed through the overproduction of amyloid-β42 (Aβ42), which triggers neurodegeneration and cognitive decline by the median ages of 44-49.
Whole-exome sequencing and whole genome sequencing revealed that this individual also carried two copies of the rare Christchurch mutation in the APOE3 allele of the APOE gene (the major susceptibility gene for late-onset Alzheimer’s disease). There were no others in the kindred that were homozygote carriers of the APOE3ch mutation, leading to the postulation that APOE3ch homozygosity is required to postpone the clinical onset of ADAD.
As a result, the functional consequences of the APOE3ch variant were characterized. It has been suggested that binding of APOE to heparin sulfate proteoglycans (HSPGs) is required to promote amyloid-β aggregation (a clinical indicator of ADAD). The team of researchers demonstrated that the APOE3ch variant displayed the lowest HSPG binding ability, and that its R136S mutation may contribute to the observed reduction in binding.
A panel of monoclonal antibodies (mAbs) was generated against a peptide that included the R136S mutation of APOE3ch. From this panel, a specific mAb (1343A) was identified to reduce the binding of wild-type APOE3 to HSPGs, suggesting that antibodies or other molecules that bind to this region may reproduce the protective effect of APOE3ch. The identity of this critical mAb (1343A) was confirmed using Rapid Novor’s REmAb® de novo sequencing service.
De novo protein sequencing provided the research team with insurance by securing the complete amino acid sequence of a therapeutic mAb candidate for ADAD. This mass spectrometry-based protein sequencing technique can be used to obtain the sequence information of any antibody or protein for biomarker discovery, characterization, and validation. Access to this structural information only broadens our understanding of disease pathogenesis and fosters the development of innovative therapeutic or preventative treatments.
Graphical Abstract
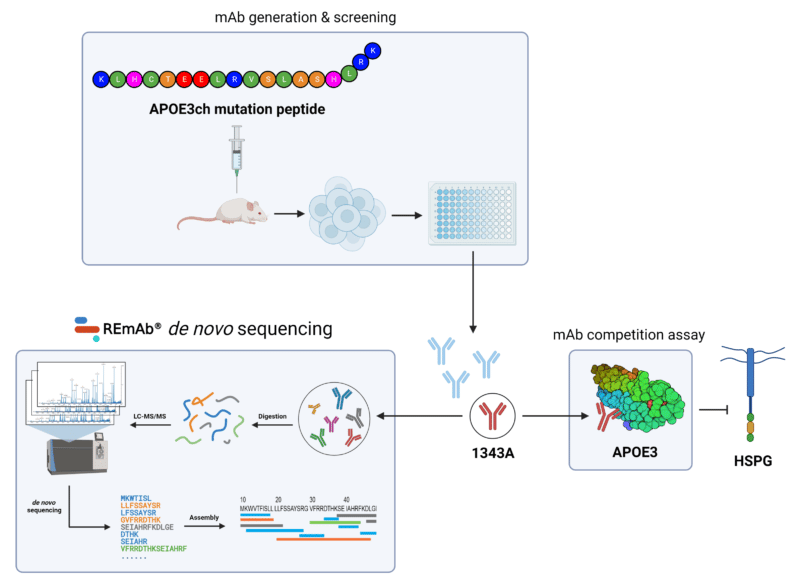
Schematic of the antibody generation campaign against APOE3. REmAb® de novo sequencing was leveraged to derive the primary amino acid sequence of 1343A, providing the security and assurance for this mAb candidate with therapeutic potential.
Key Takeaways
- A team of researchers at Harvard Medical School and University of Arizona leveraged de novo protein sequencing in the discovery and characterization of the APOE3ch biomarker and secured the sequence of a novel therapeutic mAb candidate for ADAD.
- Obtaining the primary amino acid sequence via de novo protein sequencing provides insurance of critical reagents.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

