Reviewed by María Gerpe, PhD.

Written by Yuning Wang, PhD.
July 5, 2021
Introduction
Amino acid sequencing is the process of identifying the arrangement of amino acids in proteins and peptides. Numerous distinct amino acids have been discovered in nature but all proteins in the human body are comprised of just twenty different types. Yet these few organic molecules can attach to one another in complex three-dimensional structures of near-limitless structural varieties. This underlies the immense functional diversity of proteins—and suggests the inherent value of amino acid sequencing.
Understanding even the partial sequence of amino acids in a polypeptide chain can yield valuable insights into the identity of a protein or peptide, and can help characterize its post-translational modifications.
So, how do you carry out amino acid sequencing? There are three main amino acid sequencing methods, but before we explore those in more depth, let us briefly discuss how to identify the unordered amino acid composition of a polypeptide.
How to Determine Amino Acid Composition
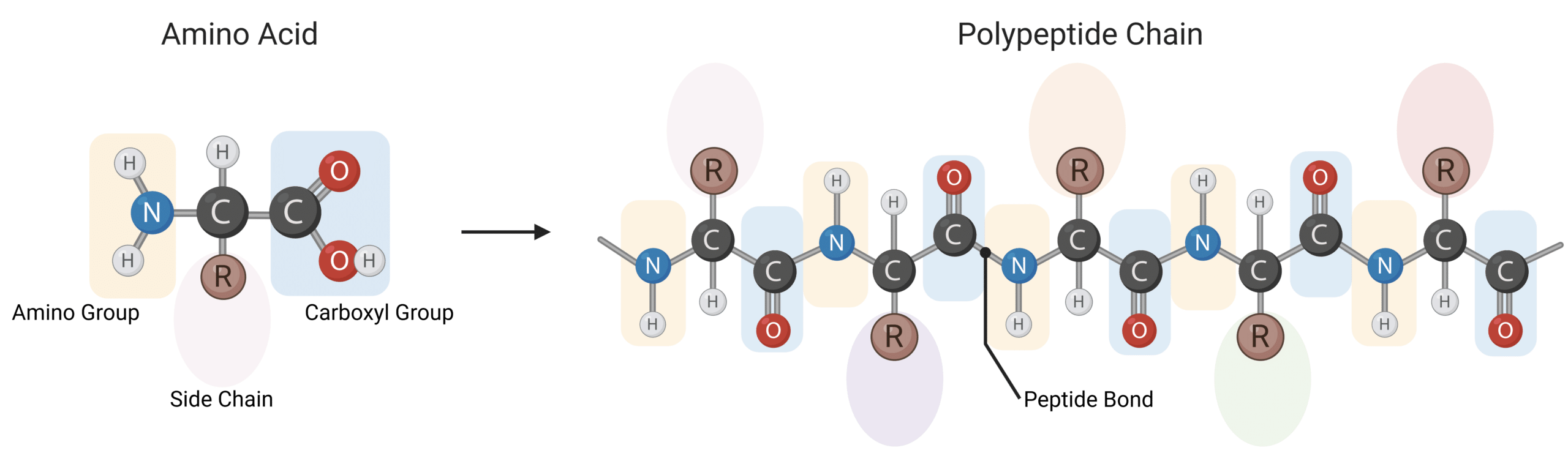
Amino acids belong to a class of organic molecules consisting of a basic amino group, an acidic carboxyl group, an organic R group—or side chain—and a central carbon atom. Abbreviated from alpha-amino carboxylic acid, amino acids are linked together by peptide bonds linking amino acids into extensive linear chains. Peptide bonds are covalent chemical bonds between the carboxyl group of one amino acid and the amino group of another (Figure 1).
Before carrying out amino acid sequencing, it is often useful to determine the unordered composition of a protein by hydrolytically degrading it then derivatizing the sample to make it more volatile and less reactive, thus increasing its suitability for analysis via ion-exchange chromatography.
The benefit of determining this unordered composition prior to amino acid sequencing is that it can help in identifying errors and may elucidate ambiguous results. It may also offer insights into the right protease to use for protein digestion.
Figure 1: Illustration of the structure of an amino acid and its polymerization into a polypeptide chain.
How to Write an Amino Acid Sequence
The primary protein structure typically begins at an amine-terminus, or N-terminus; an amino acid residue with an amine group attached to the alpha-carbon. At the other end of the primary structure is the C-terminus with an unbound carboxyl group. Each of the amino acids found in nature are represented by a single or three-letter code. For instance, alanine is expressed as Ala, or A (Figure 2). The entire sequence of protein can subsequently be notated as a string of letters from left-to-right, corresponding to order of amino acids from terminal-to-terminal.
Figure 2: Illustration of the notation used to represent amino acids in a polypeptide sequence.
Amino Acid Sequencing Methods
As mentioned, there are two main methods of amino acid sequencing: mass spectrometry and Edman degradation with a protein sequenator.
Automated Edman amino acid sequencers are offer convenient analysis of polypeptides of up to 50 amino acids long. This process is generally characterized by seven steps:
- Break apart disulphide bridges in the protein with a reducing agent
- Separate the protein complex and purify the chain(s)
- Determine the amino acid composition and terminal AAs per chain
- Fragment each polypeptide chain
- Recreate the AA sequence using these fragments
- Repeat with different fragment patterns to mitigate errors
Identification via mass spectrometry is increasingly preferred as it overcomes many of the established limitations of Edman degradation. But there are various techniques within protein mass spectrometry that make amino acid sequencing via MS techniques harder to define in brief.
De Novo Amino Acid Sequencing with Mass Spectrometry
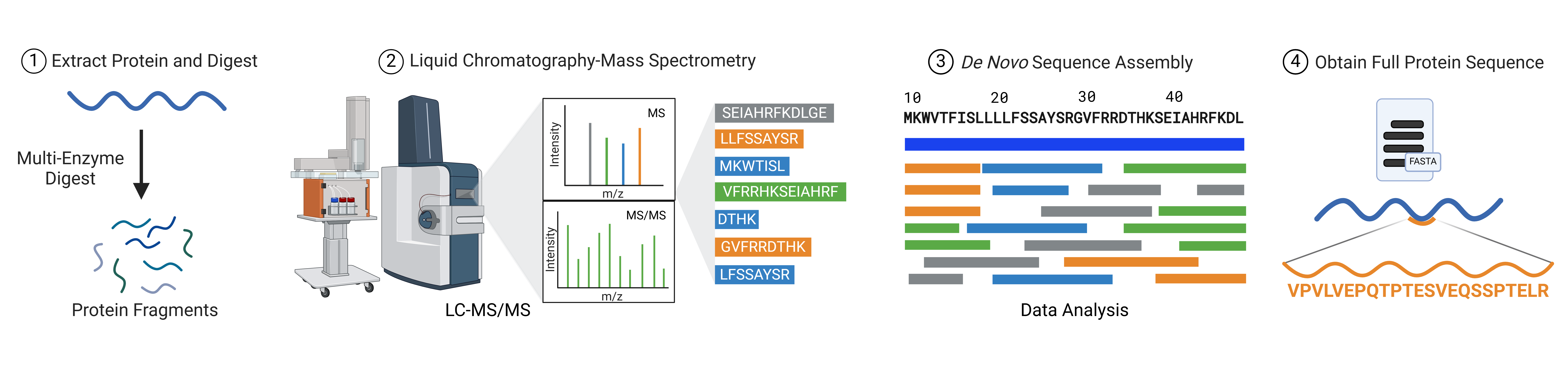
MS-based amino acid sequencing can be done with or without reference to a database of known sequences. When a database or reference sequence is used, this is called protein identification, peptide sequence identification or peptide mapping. In de novo peptide sequencing a the amino acid sequence of a peptide is determined via tandem mass spectrometry combined with bioinformatics algorithms, without a reference sequence or database. De novo protein sequencing compiles multiple overlapping de novo peptide sequences to derive a full length protein sequence. The primary benefit of de novo sequencing over conventional MS-based sequence analysis is that it allows researchers to study proteins and peptides for which there is no reference sequence – antibodies for instance. Advanced techniques including the use of multiple proteases, alternative fragmentation methods, liquid chromatography methods, high-resolution instruments and machine-learning algorithms allow rapid and highly accurate analysis of sequences and post-translational modifications (Figure 3).
Figure 3: Workflow of de novo protein sequencing using tandem mass spectrometry combined with bioinformatics algorithms.
De novo Protein Sequencing with Rapid Novor
Interested in learning more about how our antibody sequencing service can be used to determine the sequence of unknown antibodies, or how to easily distinguish between isoleucine and leucine using a novel fragmentation method? Contact a member of the Rapid Novor team today.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics