Jo, M. H., Li, J., Jaumouille, V., Hao, Y., Coppola, J., Yan, J., Waterman, C. M., Spriner, T. A., Ha, T. (2022). Single-molecule characterization of subtype-specific β1 integrin mechanics. Nat Commun 13, 7471. https://doi.org/10.1038/s41467-022-35173-w.
Abstract
Many subtypes of integrins have distinct physiological roles (ie. mediating cell adhesion, migration, and formation of tissues) primarily through integrating the intracellular and extracellular environments. The extracellular domains of integrins bind to ligands on the surface of other cells, while the cytoplasmic domains bind to adaptor proteins associated with the cytoskeleton. Upon ligand binding, integrins exist in their extended-open states, allowing the tensile force to stabilize the association between the integrins and the cytoskeletal components.
The force exerted by a single integrin on a ligand has thus far only been measured for RGD-binding integrins using fluorescent reporter systems, such as peptide-based tension sensors, DNA hairpin-based digital tension sensors, and DNA double helix-based rupturable tension gauge tethers (TGTs). These studies have demonstrated a tension threshold of over ~40pN, which is required for cell spreading by RGD-binding integrins.
The Springer Lab of Harvard Medical School and the Ha Lab of Johns Hopkins School of Medicine show that the integrin subtype α4β1 requires a notably different tension threshold of less than 12pN to activate cell spreading, compared to the threshold for RGD-binding integrins.
To address the lack of single molecule studies of force exertion by other integrin subtypes, researchers from these two groups have utilized various techniques to measure and visualize the integrin subtypes’ conformational states and tension thresholds. Among these techniques include generating Fabs conjugated to fluorescent dyes for live cell imaging and immunostaining. To generate these fluorescently conjugated Fab reagents, the Springer Lab obtained the protein sequence of the 13C2 antibody using Rapid Novor’s REmAb® de novo antibody sequencing service. Working with the exact amino acid sequence obtained through de novo protein sequencing can support the development of novel antibody-based reagents, ensuring accurate and reliable tools for molecular imaging.
Graphical Abstract
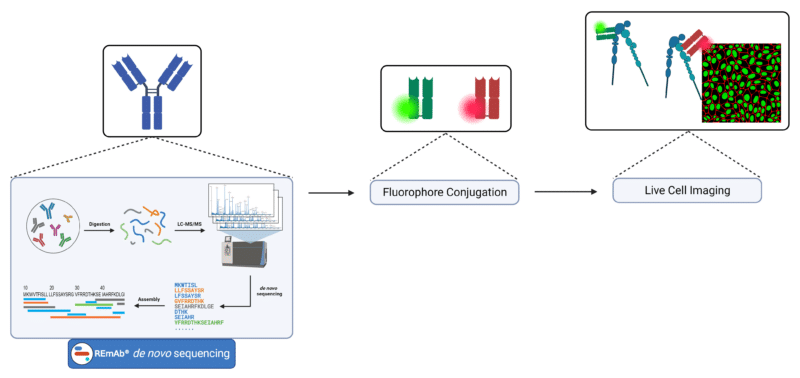
Schematic for the generation of fluorescently conjugated Fab reagents for imaging applications.
Key Takeaways
- The Springer Lab and Ha Lab applied various techniques for the single molecule analysis of force exertion by other integrin subtypes to activate cell spreading.
- Fluorescently conjugated 13C2 Fab reagents were generated for the visualization of the conformational states of the integrin subtype α4β1, which was demonstrated to have a tension threshold of 12pN.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

