August 10, 2021

1Rapid Novor, Inc, Kitchener, Ontario, Canada
Abstract
The veterinary field has witnessed many important advances in the past few decades; however, the lack of immunological reagents continues to hamper progress in veterinary research and at the clinic. This shortage in immunological reagents to study and monitor diseases impacts not just animals, but humans. The rise in frequency of emerging zoonotic diseases and the current pandemic are important reminders of this dire need. Lack of antibody standards also impact antimicrobial resistance surveillance, and companion animal health. Though antibodies are often discovered via nucleotide sequencing means, next generation protein sequencing offers a faster and more comprehensive approach to discover antibodies no matter the species or isotype.
Key Takeaways
Protein sequencing offers the following advantages:
- No culling of producer animals
- Direct access to the protein, especially useful when no or scant DNA
- Provides information on PTMs, and additional light and heavy chains of key circulating clones
- Can fill in gaps of NGS transcripts
- Complements existing nucleotide sequencing pipelines to captures a more complete picture of the humoral immune response
- Can be used for disease monitoring
State of Veterinary Medicine and Research – a short primer
Since 2006, the One Health Initiative (OHI)’s goal has been to demonstrate the inextricable link between humans, animals, and the environment2. Certainly, the current global pandemic is a great testament to the ties between climate change, humans, and animals that OHI has been working to highlight. The rise of other zoonotic diseases (e.g., Hendra, and Nipah viruses) not only directly affect humans through disease transmission but may also result in deep impacts to the food supply3. Likewise, the rise of antibiotic resistance is a great threat affecting both human and animal lives3. Policies have been put in place since the 2000s by the FDA and Health Canada to curve antibiotic overuse especially in farming practices4,5.
Moreover, as human and animal populations continue growing worldwide, the demand for products to sustain them also rises, and with it the need for waste management and toxicology surveillance. The latter becomes ever pressing in animal agriculture as production animals are often the first to show symptoms from toxin exposure6. Finally, developed countries have seen the lifespan of companion animals significantly increase in the last decade alone, and the trend is expected to continue. With a lengthier lifespan, the propensity for neoplastic diseases and comorbidities also rises. Because quality of owner care has drastically improved, there is demand for better therapeutics and diagnostics, particularly those of non-invasive nature.
However, the lack of commercially available and unvalidated antibody reagents across many species continues to undermine progress in veterinary research7. The lack of immunological reagents is mainly due to a limited understanding of other species’ immune systems7,8, although the very lack of antibodies for immunoassays and research also hampers the study of the adaptive immune system. The lack of whole genome sequencing data for many is also a contributing factor to the crisis in immunoreagent availability. To solve this and other issues, an initiative was founded in the early 2000s with the goal of decoding species’ proteomes: the Human Proteome Project Organization (HUPO)9. Proteomics, and in particular protein sequencing, are promising tools that can be exploited to:
- Control zoonotic infections (e.g., therapeutics, diagnostics, and vaccine development)
- Improve food safety (e.g., antimicrobial resistance)
- Advance Veterinary Research and Medicine (e.g., companion animal health)
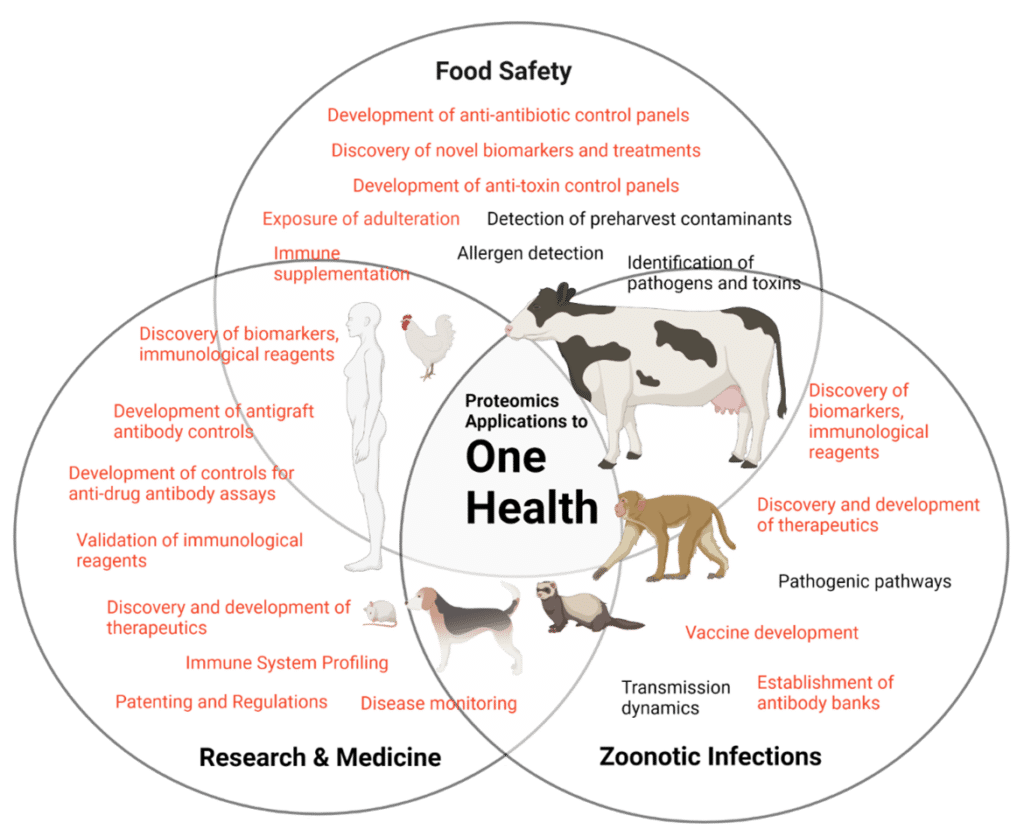
Figure 1. Proteomics applications in veterinary medicine especially as it applies to the One Health Initiative, which includes food safety (e.g., antibiotic resistance, toxins, and other chemicals), research and medicine (e.g., therapeutics, diagnostics, disease monitoring, and translational research), and zoonotic infections control. Figure adapted from Katsarou et al. (2021) In orange: applications that can also be carried out via protein sequencing.
Zoonotic Infection Control
Over a billion people worldwide depend on livestock for employment, and billions more rely on animals for food. The economic value of animal agriculture surpassed $180 billion USD last year10. However, disease outbreaks highly impact this growth; emerging zoonotic diseases11, in particular, are a constant threat to the health of both humans and animals. Examples of such diseases include the yearly flu, Hendra and Nipah viruses, and vector-based diseases (e.g., malaria and Lyme disease). The cost of only one zoonotic disease, Foot-and-Mouth disease (FMDV), from a small outbreak, in cattle alone, would cost Canada $2 billion over 5 years12. As such, disease prevention in animal agriculture is key.
Prevention and Treatment
Other than the highly contagious FMDV, coronavirus infections can cause disease in both mammals and birds, specifically cattle, pigs, and chickens13. Though capable of infecting more than one species, none of the coronaviruses capable of infecting cattle, pigs, and chickens have been shown to be transmissible to humans13. Nevertheless, the current pandemic remains a cautionary tale for zoonoses14, and highlights the importance of immunological reagents in monitoring, and thus preventing outbreaks.
Furthermore, some zoonotic diseases such as FMDV remain elusive to vaccine development due to its high antigenic diversity and few neutralization studies in natural hosts15. As such, antibody banks have been developed worldwide to combat outbreaks at the ready. The latter also proving the dire need for antibody discovery in veterinary medicine.
Research
Aside from viral diseases, there are also zoonotic prion and bacterial transmissions that can be devastating to animals, and the animal agriculture industry and large16,17. Perhaps the most commonly known is the one colloquially referred to as mad cow disease, which burdened the health of animals, and resulted in a few human cases in the UK in the late 1990s18. Mad cow disease is caused by prions – proteins, which are abnormally folded, highly prone to aggregation, and extremely resistant to degradation17,18.
Because protein-protein interactions are integral to the pathogenesis of prion diseases, proteomics is needed to understand their mechanism of action, and novel methods may be needed to identify the interactions17. Antibodies can help identify these interactions and even aid in the structural characterization of proteins. A recent study utilized an antibody as a purification tool for a difficult-to-purify protein complex prior to CryoEM structural analysis19.
Food Safety
Antimicrobial resistance
As mentioned before, some zoonotic diseases include diseases caused by bacteria. The latter Mycobacterium bovis, the most commonly transmitted tuberculosis agent in cattle17. However, it also causes disease in humans early and globally. Tuberculosis is one of the bacterial diseases associated with superbugs20.
Superbugs or the global threat of antimicrobial resistance is a serious issue affecting animal agriculture and humans. In the past, antibiotics were used to supplement feeds for cattle, and other production animals21. In fact, 80% of all antibiotic use is reserved for animal agriculture21. Antibiotics are divided into different classes depending on their importance to human health22. Some antibiotics such as tetracycline, are known as class III antibiotics because they have few potential replacements in human clinical treatment23. For this reason, since 2013, the FDA, and later on the CFIA began regulating the use of antibiotics at farms, and also testing food sources for the presence of high classes of antibiotics, like tetracycline24.
At Rapid Novor, using de novo antibody protein sequencing, we have decoded anti-antibiotic antibodies such as anti-tetracyclines to be used in the development of diagnostics to test for their presence in foods. With the rise of superbugs, it is now vital to ensure, antibiotics do not cross over into our food supply25, and further increase resistance.
Toxin and chemical surveillance
Another important aspect of OHI is the collaboration among veterinarians, physicians and toxicologists. This is because many toxin or chemicals in the environment have first resulted in illnesses in animal herds6. The most pervasive example is perhaps DDT; its use was still debated in the early 2010s with regards to malaria prevention 26.
To date, antibody therapy remains the only effective treatment against toxins27. In fact, the field of immunotoxicology has been an important source of cancer and infection research since the 1970s, given that exposure to toxins often results in an increased propensity to diseases such as cancer28. As such, because the immune system is such an integral part of how humans respond to the environment, immunological reagents are essential to the toxicological research toolbox.
Companion Animals
The lifespan of companion animals has significantly increased over the past decades due to advances in research and improved care by pet owners29. With a greater lifespan, autoimmune complications like arthritis and cancers such as multiple myeloma, breast and lung cancers have also increased. Each year, 10% of dogs are diagnosed with cancer in the US alone30. Earlier diagnosis often correlates with better prognosis. As such, there’s demand for better diagnostics and therapeutics for companion animals. However, the lack of immunoreagents and monoclonal antibody (mAb) therapeutics continues to hinder progress in companion animal medicine, research, and their quality of life.
Liquid Biopsies for Improved Detection
The lack of immunoreagents also implies that there is a lack in diagnostic tests for many companion animal diseases like cancer30. For some, as is the case for multiple myeloma (MM), diagnostic tests are available, but they suffer from poor sensitivity and specificity. Despite being rare in companion animals, MM is the second most diagnosed neoplastic condition in dogs; it is also diagnosed in cats and horses31. The most common way to treat companion animals for MM is with chemotherapeutics routinely used to treat human MM; though they have been shown to reduce tumor size in dogs, they do not dramatically improve prognosis, which is already poor in companion animals with MM.
Though immunotherapy has greatly improved the prognosis of humans with MM32, therapeutic mAbs have yet to be used for MM in companion animals31. The lack of understanding of their immune system, and few sensitive and specific diagnostics may be to blame30. Because most MM cases involve the secretion of an aberrant monoclonal antibody, termed the M-protein, proteomics in this case can be a powerful tool in comparison to genomics or transcriptomics. Proteomics is ideally suited to identify more sensitive methods for screening, and more aggressive and direct therapeutics to treat MM33.
Anti-Drug Antibody Assays
In 2017, the first mAb therapeutic for animal use, developed by Zoetis, was approved for the treatment of atopic dermatitis34. Still, in contrast to humans that had over 70 therapeutic mAbs approved by the FDA in 2019 alone, companion animals, namely dogs, have but a handful35,36. As mentioned before, a key culprit is the lack of understanding of the immune system of many animals, except for mice and humans30. Knowledge of the factors within the immune system that result in immunogenicity are indispensable to therapeutic development. Specifically, autoantibodies against IgG, the backbone of many therapeutic mAbs, can hamper their efficacy and safety.
Autoantibodies – rheumatoid factors, anti-Fab and anti F(ab’)2 autoantibodies, and anti-idiotypic antibodies – can impact a drug’s efficacy directly or indirectly, by obstructing binding with target through paratope interaction (anti-idiotypic), or by affecting stability (anti-Fab and anti F(ab’)2 autoantibodies) or occluding target epitopes (rheumatoid factors)35. ADAs are ligand-based assays that test for immunogenicity or the presence of autoantibodies that could prevent the proper function of therapeutics37.
ADA assays rely on antibody standards meant to be representative of the target animals’ autoantibodies. Currently, researchers utilize the target species’ purified serum or mAb panels obtained via nucleotide sequencing technologies to develop ADA standards. But, as previously noted, these technologies often require culling of the producer animal, or typically fail to capture the total circulating antibody repertoire. On the other hand, the target animal’s freely circulating antibody proteins would better reflect the autoantibodies impacting drug efficacy, and safety.
Current Methods in Developing Immunological Reagents, Diagnostics and Therapeutics
Hybridoma development, next generation sequencing of B cells, and display technologies are traditional approaches primarily used to develop antibodies for therapeutics38-40. However, these methods have many shortcomings that result in a lengthier process of discovery and development.
Other than being often lost or misplaced, hybridoma cell lines often yield mAbs with aberrations, or with additional chains, resulting in loss of specificity and/or cross-reactivity41-43. In next generation sequencing of B-cells, important clones, or pairing information may be missing, resulting in the discovery of antibodies that are not truly representative of the circulating antibody repertoire39,40,44,45. Furthermore, NGS-based mAb discovery in animals requires spleen collection and thus, the death of the producer animal46.
Display technology is dependent on nucleotide sequencing approaches to generate synthetic repertoire libraries for subsequent affinity maturation. As such, it is subject to the same limitations of nucleotide sequencing approaches in addition to not having post-translational modifications (PTMs) (e.g., glycans) required for stability47. Not truly capturing the natural antibody repertoire results in prolonged target validation, delaying use at the clinic.
For diagnostics, and reagent development, hybridoma technologies, and collection of animal sera are the most used methods to date to develop antibodies. The former is used to produce mAbs, and the latter to generate polyclonal antibodies (pAb). However, these methods suffer from reproducibility-related shortcomings.
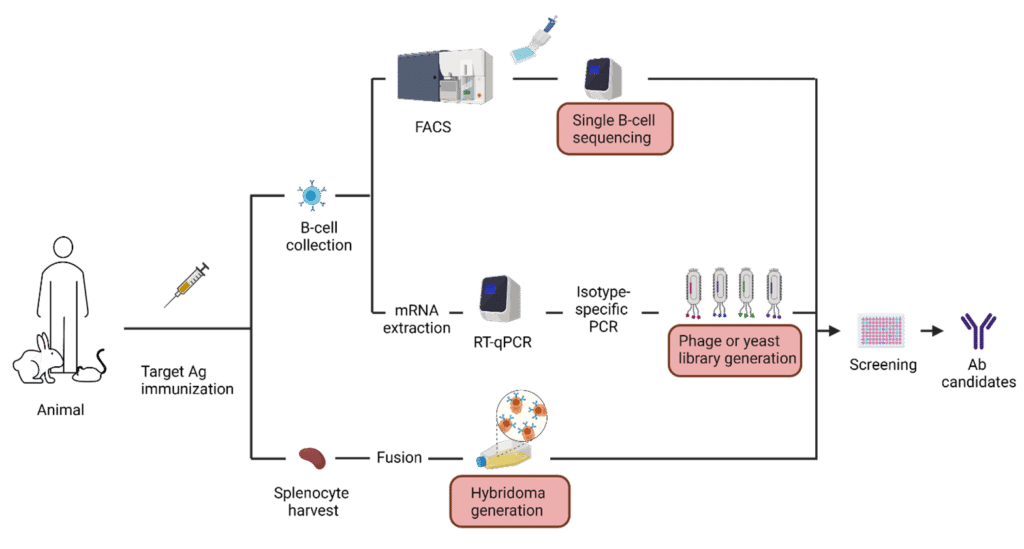
Figure 2. Infographic illustrating common methods used in therapeutic antibody development.
Current Monitoring of Immune Responses in Animals
Currently, one of the ways that disease prevention is carried out in animal agriculture is via selection of production animals with enhanced immune responses. This is achieved in dairy cattle with the High Immune Response (HIR) technology, and the Immunity+™ sire line48. However, studies in pigs have also shown that tracking the immune response is vital to understanding which animals to breed for disease prevention49.
Particularly, knowledge of the antibody-mediated immune response has proven critical to breeding animals with enhanced and appropriate immune responses. This is because antibodies are crucial against extracellular pathogens49, but they can also be used for phenotypic biomarkers of enhanced immune responses against intracellular pathogens50.
Because adaptive immune responses have been shown to be heritable51, tracking the adaptive immune system is mainly done via genetic-based assays. However, including proteomics is critical to understanding enhanced immune responses17 in production animals because:
- The use of proteomics ensures a more comprehensive systems biology approach
- With proteomics, researchers can have a greater global understanding of the immune interactome (the collection of signaling and reactions between the intrinsic, innate, and adaptive immune systems)
- Disease symptoms could be monitored through blood draw
- Proteomics can allow the discovery of new immunological reagents
Let’s look at failure of transfer of passive immunity (FTP) syndrome in calves as an example. Colostrum must be given to calves within 24 hours of being born to ensure necessary immunity is transferred from the mother because immunoglobulins are unable to pass through the bovine placenta52. Immune factors transferred through the first milk – colostrum – include antibodies, cytokines and others; of these, antibodies are perhaps most important. FTP occurs when the colostrum a calf is fed does not adequately protect the naïve calf from its new environment52. Profiling the components of the colostrum given to calves could elucidate the makeup of the antibody repertoire that calves first access from their mothers, and in the event of FTP occurrence, supplementation with the antibodies most needed by calves would ensure survival.
However, FTP is not the only potential application; mastitis, which is also often treated with a combination of medications, including immunotherapy53 could benefit from mAb cocktails that could be discovered through protein sequencing. As part of the proteomics arsenal, next generation protein sequencing can be used to sequence the antibodies and T-cell receptor proteins to directly access the immune response output, without relying on cellular input.
Protein Sequencing as a Complementary and Comprehensive Solution to Address Reagent, Biomarker, and Therapeutic Needs in Veterinary Medicine and Research
The process of translation does not result in one gene outputting one protein. A commonly used example is the human genome; humans’ 20 000 or so genes are translated into millions of proteoforms due to PTMs, alternative splicing, and germline variants. Certainly, to understand the function of proteins, knowledge of their primary structure – the amino acid sequence and PTMs – is critical. But finding the PTMs is not the only challenge in decoding proteins; nucleotide sequence information is often missing or scant in research due to lost cell lines, little cellular input, or degraded or low concentration of DNA43,54. In fact, the latter two are frequently given reasons for using de novo protein sequencing technology55.
For these reasons, MS-based protein sequencing remains a relevant tool to unearth the primary structure of proteins, especially since nucleotide sequencing tools are simply blind to PTMs. Because antibodies are by nature very diverse, and because they harbor PTMs (e.g., glycans) that affect binding, stability, and half-life56,57, protein sequencing is particularly well suited to decoding them. Protein sequencing can also be applied to existing pipelines to complement existing nucleotide sequencing efforts, to capture a more complete picture of antibody structure and function1,45,58.
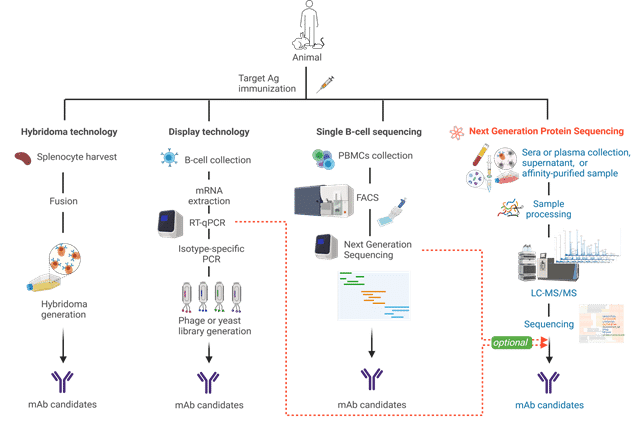
Figure 4. Illustration comparing different antibody discovery technologies.
Antibodies are crucial to mechanistic studies and biomarker identification. They’re also critical components in major tools used in the clinic and at the bench (e.g., ELISAs, and immunohistochemistry7). But lack of species relevant antibodies with which to conduct a myriad of assays, and lack of proper validation threatens medical and research advances. To understand loss of function problems, mass spectrometry-based proteomics and protein sequencing is required59-64.
EasyMTM: Monitoring Multiple Myeloma Sensitively and Specifically
EasyMTM is a mass spectrometry-based, non-invasive test that monitors the presence of the M-protein, a widely used biomarker for diagnosing MM33,65. Our test has been validated in humans through two retrospective clinical trials with two major cancer centers in North America.
Because our test targets the protein sequence, and our protein sequencing technology can sequence any species, EasyMTM may be applicable to other animals with MM, such as dogs, cats, and horses.
Our test has additional advantages. Because we can sequence the M-protein, new and more direct therapeutics could be developed from this information. Furthermore, since it focuses on the sequence of the antibody protein, the concept could be repurposed to other autoimmune diseases.
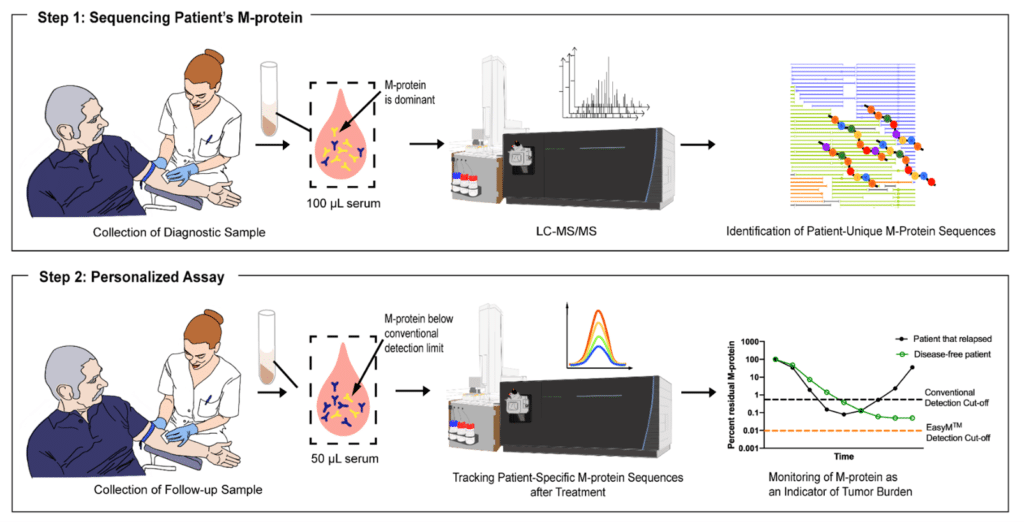
Figure 3. EasyMTM is the first test employing protein sequencing to monitor the M protein of Multiple Myeloma. Firstly, a sample is collected non-invasively (serum) from the patient, and their M-protein is sequenced to identify patient-specific unique peptides (Step 1) that are then tracked in future collected samples (Step 2) to predict M-protein earlier than conventional means such as SPEP and IFE.
Advantages of Protein Sequencing over Nucleotide Sequencing
De novo protein sequencing proves to be an excellent tool when there is limited or no nucleic material. Plus, where NGS is routinely conducted, it greatly complements antibody discovery pipelines. De novo protein sequencing does not require culling the producer animal as it can be done directly on the protein purified from a blood sample. Furthermore, it includes bioinformatics analysis that permits lineage studies. Most importantly, it directly accesses the protein by extracting the sequence and PTMs important for function and structure. As such, protein sequencing truly captures a more complete picture of the humoral immune response to inform therapeutics, diagnostics, and reagent development.
Though it can help when nucleic material is limited, or with new species, NGPS does not aim to replace nucleotide sequencing efforts. Certainly, it enhances nucleotide sequencing strategies to achieve a truly global understanding of the antibody leads destined for the clinic and research. To complement efforts in existing pipelines and characterize the broad BCR, de novo protein sequencing can be applied either as a proteomics-only or as a proteogenomics approach at different stages of the antibody discovery process, depending on client needs and project complexity.
Why is protein sequencing not widely used?
Efforts to decode proteomes are ongoing (e.g., HUPO), but this task has proven difficult. Though protein sequencing technology has greatly advanced, both academic and industry labs do not currently effortlessly sync two key technologies: MS-based protein sequencing and machine learning-driven bioinformatics.
It is not so difficult to get up to 95% coverage of the protein sequence — especially when one has a similar protein to start with. However, the remaining 5% is usually highly variable, and therefore can rarely be determined by simple local alignment based on known protein sequences. Software adapted from whole proteomics approach can underperform in these areas. Good data analysis starts with good raw data generation; therefore, a laboratory specializing in de novo protein sequencing using mass spectrometry and specifically de novo antibody sequencing using high resolution mass spectrometry instrumentation is also crucial.
De novo protein sequencing is a robust tool to discover biomarkers, develop therapeutics, and monitor disease. Although the principle of protein sequencing is easy to understand, it requires significant expertise in both the mass spectrometry protocols and informatics data analysis to achieve the correct result.
Next Generation Protein Sequencing Enhances and Accelerates Antibody-Centric Research
Mass spectrometry tends to yield copious amounts of data, often used for database search or de novo protein sequencing. As many genomes are yet to be sequenced, the latter is the only tool capable of decrypting novel proteins. Next generation protein sequencing is a term used to coin the advances in mass spectrometry-based elucidation of the amino acid sequence of proteins, possible through integration with artificial intelligence to yield de novo protein sequencing.
From studying ancient remains2 to protein characterization, and from development of therapeutics37 to disease monitoring16, protein sequencing remains an important weapon in the armamentarium needed to understand biological processes and treat diseases. In 2019, a study in nature showed that protein sequencing was vital to confirming the ancestry of ancient Denisovan remains when DNA information was missing or scant. Since its founding in 2015, Rapid Novor has helped discover checkpoint inhibitors, characterize the structure of several proteins95, develop monoclonal therapies87, establish patentable research48, and monitor disease using protein sequencing.
Applications of Protein Sequencing
Therapeutics Development
Knowledge of the antibody three-dimensional structure is critical to guide rational drug design. Recently, Rapid Novor’s de novo protein sequencing services facilitated the structural characterization of two potent henipavirus anti-glycoprotein mAbs32 without relying on cell line, genomics or transcriptomics data. Our de novo protein sequencing platform has also helped identify novel and natural anti-checkpoint inhibitor antibodies33 and safeguarded the discovery of bispecific therapeutics from one of the top biopharmaceutical companies globally34.
Diagnostics Development
Because protein sequencing can directly access the antibody proteins from protein-rich samples, we were able to extract several antibodies from a polyclonal mixture for a client. In turn, we became the first in industry to do this using only proteomics in barely two weeks – an unprecedented task35.
Reagents Development
Our clients also routinely use our protein sequencing services to develop reagents for commercial use, and for in-house process development.
Protein Characterization
Our de novo antibody protein sequencing technology continues to be utilized in the field of structural biology. Protein sequences obtained via our protein sequencing technology have helped researchers examine protein structures and drug-target interactions at the atomic level, defining directions for developing therapeutics against various diseases36,37.
Patenting and Other Regulations
Our technology is often used to protect newly discovered biologics, including bispecific34, and immunomodulatory antibodies38. Moreover, it is becoming increasingly used to generate anti-drug antibodies (ADAs), control reagents in assays testing the clinical efficacy and safety of a biological drug39. Such antibodies mimic the natural ADA response to the biological drug. These positive controls typically consist of animal-derived (e.g., rabbits) pooled polyclonal antibodies (pAb) or human monoclonal antibody (mAb) reference panels against the target protein drug39. As such, the specific sequence is critical to the development of ADA assays, and in many cases, regulatory paperwork may require their exact sequence in a short turnaround (within six months).
De novo Protein Sequencing at Rapid Novor
Workflow
An aqueous or lyophilized sample, or supernatant containing 100 µg of protein (~80% purity) from any species or any isotype – in the case of antibodies is first processed via purification, if needed. A multi-enzyme or protease digest is then performed prior to liquid chromatography tandem mass spectrometry analysis (LC-MS/MS). Sequencing and assembly are done in parallel to MS analysis. Machine learning algorithms and a team of bioinformatics specialists proof check the assembled sequences. A report containing notable observations such as PTMs along with the sequence is prepared and sent right to the client’s inbox within days of the sequencing experiment, depending on the amount of antibodies.
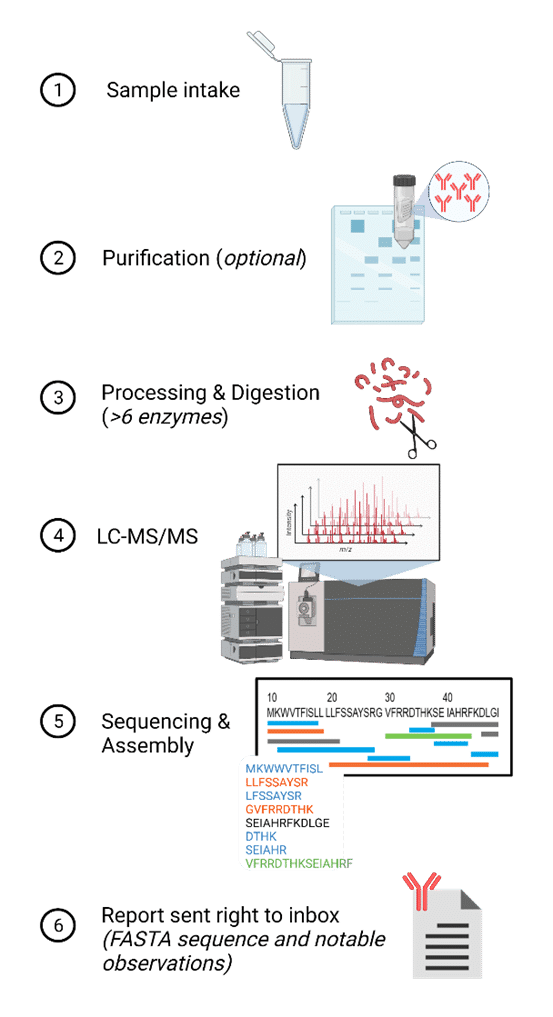
Figure 5. Illustration depicting an overview of the protein sequencing workflow at Rapid Novor.
Deciding between DNA and Protein Sequencing
Protein sequencing is not a replacement technology. Rather, it enhances the study of protein targets. Certainly, protein sequencing is a great complementary tool to pipelines that already rely on nucleotide sequencing. In the case of lost cell lines, cell lines behaving erratically, or limited nucleic material, it is a great substitute to DNA sequencing. However, for other projects aiming to characterize protein targets, or further engineer and develop antibody leads, protein sequencing can be used alongside nucleotide sequencing tools for a more comprehensive approach with less hiccups at the clinical stage.
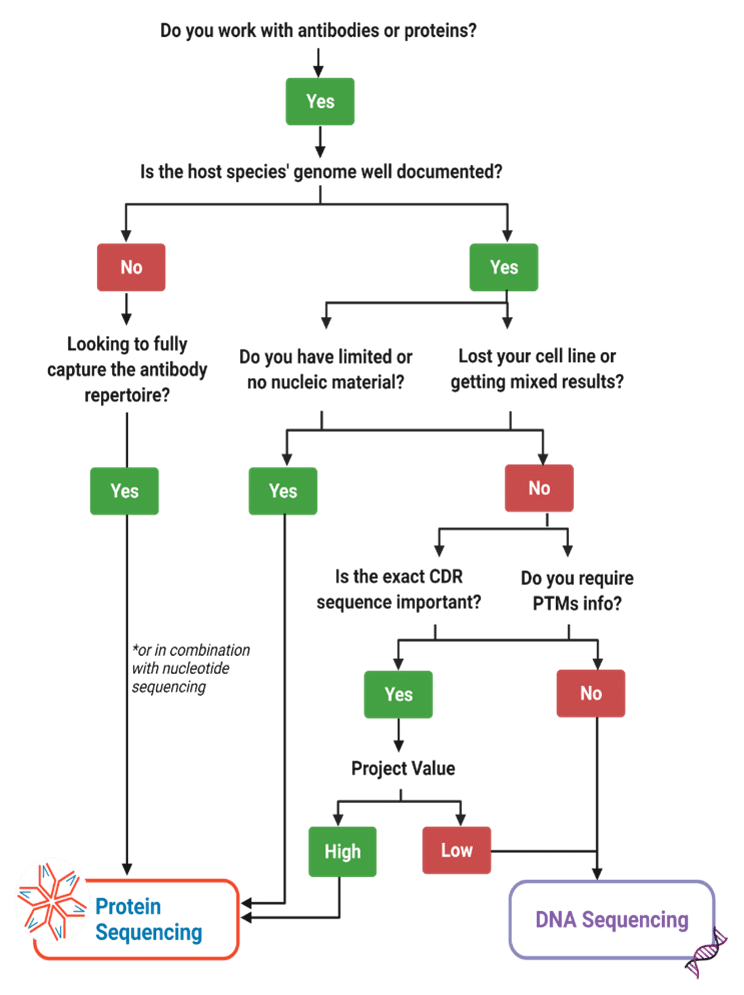
Figure 6. Decision matrix tool to decide how to best approach a project’s assessment of a protein target.
Conclusions
The biggest obstacle in advancing veterinary medicine and research continues to be a lack of immunological reagents to better understand the immune system of different species, and to develop solutions to antimicrobial resistance, engineer better treatments and diagnostics for companion animals, and curb the threat of emerging zoonotic diseases.
Protein sequencing remains a staple of antibody discovery, particularly because of its ability to access the PTMs, in contrast to nucleotide sequencing methodologies. As such, it is a great tool to populate the immunological toolbox and expand the knowledge of the yet-to-be-elucidated immune system of many species.
References
1. Aebersold, R. et al. How many human proteoforms are there? Nat Chem Biol 14, 206-214, doi:10.1038/nchembio.2576 (2018).
2. Sousa, E. et al. Primary sequence determination of a monoclonal antibody against α-synuclein using a novel mass spectrometry-based approach. International Journal of Mass Spectrometry 312, 61-69, doi:10.1016/j.ijms.2011.05.005 (2012).
3. Rapid Novor, I. Hybridoma Loss Prevention, <https://www.rapidnovor.com/hybridoma-loss-prevention/> (2021).
4. Xin, H. & Cutler, J. E. Hybridoma passage in vitro may result in reduced ability of antimannan antibody to protect against disseminated candidiasis. Infect Immun 74, 4310-4321, doi:10.1128/IAI.00234-06 (2006).
5. Bradbury, A. R. M. et al. When monoclonal antibodies are not monospecific: Hybridomas frequently express additional functional variable regions. MAbs 10, 539-546, doi:10.1080/19420862.2018.1445456 (2018).
6. Chen, F. et al. A late Middle Pleistocene Denisovan mandible from the Tibetan Plateau. Nature 569, 409-412, doi:10.1038/s41586-019-1139-x (2019).
7. Hughes, C., Ma, B. & Lajoie, G. A. De novo sequencing methods in proteomics. Methods Mol Biol 604, 105-121, doi:10.1007/978-1-60761-444-9_8 (2010).
8. Jefferis, R. & Lefranc, M. P. Human immunoglobulin allotypes: possible implications for immunogenicity. MAbs 1, 332-338, doi:10.4161/mabs.1.4.9122 (2009).
9. Kaur, H. Characterization of glycosylation in monoclonal antibodies and its importance in therapeutic antibody development. Crit Rev Biotechnol 41, 300-315, doi:10.1080/07388551.2020.1869684 (2021).
10. Jovčevska, I. & Muyldermans, S. The Therapeutic Potential of Nanobodies. BioDrugs 34, 11-26, doi:10.1007/s40259-019-00392-z (2020).
11. Zaroff, S. & Tan, G. Hybridoma technology: the preferred method for monoclonal antibody generation for. Biotechniques 67, 90-92, doi:10.2144/btn-2019-0054 (2019).
12. Alfaro, J. A. et al. The emerging landscape of single-molecule protein sequencing technologies. Nat Methods 18, 604-617, doi:10.1038/s41592-021-01143-1 (2021).
13. Toleikis, L. & Frenzel, A. Cloning single-chain antibody fragments (ScFv) from hyrbidoma cells. Methods Mol Biol 907, 59-71, doi:10.1007/978-1-61779-974-7_3 (2012).
14. Lu, R. M. et al. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci 27, 1, doi:10.1186/s12929-019-0592-z (2020).
15. Boyd, D. et al. Isolation and characterization of a monoclonal antibody containing an extra heavy-light chain Fab arm. MAbs 10, 346-353, doi:10.1080/19420862.2018.1438795 (2018).
16. Harris, C. et al. Identification and characterization of an IgG sequence variant with an 11 kDa heavy chain C-terminal extension using a combination of mass spectrometry and high-throughput sequencing analysis. MAbs 11, 1452-1463, doi:10.1080/19420862.2019.1667740 (2019).
17. Regl, C. et al. Dilute-and-shoot analysis of therapeutic monoclonal antibody variants in fermentation broth: a method capability study. MAbs 11, 569-582, doi:10.1080/19420862.2018.1563034 (2019).
18. Strasser, L. et al. Proteomic Profiling of IgG1 Producing CHO Cells Using LC/LC-SPS-MS. Front Bioeng Biotechnol 9, 569045, doi:10.3389/fbioe.2021.569045 (2021).
19. Thakur, A. et al. Identification, characterization and control of a sequence variant in monoclonal antibody drug product: a case study. Sci Rep 11, 13233, doi:10.1038/s41598-021-92338-1 (2021).
20. Benichou, J., Ben-Hamo, R., Louzoun, Y. & Efroni, S. Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology 135, 183-191, doi:10.1111/j.1365-2567.2011.03527.x (2012).
21. VanDuijn, M. M., Dekker, L. J., van IJcken, W. F. J., Sillevis Smitt, P. A. E. & Luider, T. M. Immune Repertoire after Immunization As Seen by Next-Generation Sequencing and Proteomics. Front Immunol 8, 1286, doi:10.3389/fimmu.2017.01286 (2017).
22. Slatko, B. E., Gardner, A. F. & Ausubel, F. M. Overview of Next-Generation Sequencing Technologies. Curr Protoc Mol Biol 122, e59, doi:10.1002/cpmb.59 (2018).
23. VanDuijn, M. M., Dekker, L. J., van, I. W. F. J., Sillevis Smitt, P. A. E. & Luider, T. M. Immune Repertoire after Immunization As Seen by Next-Generation Sequencing and Proteomics. Front Immunol 8, 1286, doi:10.3389/fimmu.2017.01286 (2017).
24. Kunert, R. & Reinhart, D. Advances in recombinant antibody manufacturing. Appl Microbiol Biotechnol 100, 3451-3461, doi:10.1007/s00253-016-7388-9 (2016).
25. Meyer, L. et al. A simplified workflow for monoclonal antibody sequencing. PLoS One 14, e0218717, doi:10.1371/journal.pone.0218717 (2019).
26. Guthals, A. et al. De Novo MS/MS Sequencing of Native Human Antibodies. J Proteome Res 16, 45-54, doi:10.1021/acs.jproteome.6b00608 (2017).
27. Gilchuk, P. et al. Proteo-Genomic Analysis Identifies Two Major Sites of Vulnerability on Ebolavirus Glycoprotein for Neutralizing Antibodies in Convalescent Human Plasma. Frontiers in Immunology 12, doi:10.3389/fimmu.2021.706757 (2021).
28. Rouet, R., Jackson, K. J. L., Langley, D. B. & Christ, D. Next-Generation Sequencing of Antibody Display Repertoires. Front Immunol 9, 118, doi:10.3389/fimmu.2018.00118 (2018).
29. Chan, C. E., Lim, A. P., MacAry, P. A. & Hanson, B. J. The role of phage display in therapeutic antibody discovery. Int Immunol 26, 649-657, doi:10.1093/intimm/dxu082 (2014).
30. Chen, G. & Sidhu, S. S. Design and generation of synthetic antibody libraries for phage display. Methods Mol Biol 1131, 113-131, doi:10.1007/978-1-62703-992-5_8 (2014).
31. Hammers, C. M. & Stanley, J. R. Antibody phage display: technique and applications. J Invest Dermatol 134, 1-5, doi:10.1038/jid.2013.521 (2014).
32. Dang, H. V. et al. Broadly neutralizing antibody cocktails targeting Nipah virus and Hendra virus fusion glycoproteins. Nat Struct Mol Biol 28, 426-434, doi:10.1038/s41594-021-00584-8 (2021).
33. Bratslavsky, G. T., I. in Developmental Therapeutics Vol. 30 v188 (Annals of Oncology, 2019).
34. Ganesan, R., Grewal, I., S. & Singh, S. MATERIALS AND METHODS FOR MODULATING T CELL MEDIATED IMMUNITY. (2020).
35. in News Medical Life Sciences (2021).
36. Nešić, D. et al. Cryo-Electron Microscopy Structure of the αIIbβ3-Abciximab Complex. Arterioscler Thromb Vasc Biol 40, 624-637, doi:10.1161/ATVBAHA.119.313671 (2020).
37. Dai, D. L. et al. Structural Characterization of Endogenous Tuberous Sclerosis Protein Complex Revealed Potential Polymeric Assembly. Biochemistry 60, 1808-1821, doi:10.1021/acs.biochem.1c00269 (2021).
38. Bernard, M., A.; & Tacahdo, S., D. COMPOSITIONS AND METHODS FOR TREATING INFLAMMATORY DISEASES. (2019).
39. Shibata, H. et al. Comparison of different immunoassay methods to detect human anti-drug antibody using the WHO erythropoietin antibody reference panel for analytes. J Immunol Methods 452, 73-77, doi:10.1016/j.jim.2017.09.009 (2018).
2. Sousa, E. et al. Primary sequence determination of a monoclonal antibody against α-synuclein using a novel mass spectrometry-based approach. International Journal of Mass Spectrometry 312, 61-69, doi:10.1016/j.ijms.2011.05.005 (2012).
3. Rapid Novor, I. Hybridoma Loss Prevention, <https://www.rapidnovor.com/hybridoma-loss-prevention/> (2021).
4. Xin, H. & Cutler, J. E. Hybridoma passage in vitro may result in reduced ability of antimannan antibody to protect against disseminated candidiasis. Infect Immun 74, 4310-4321, doi:10.1128/IAI.00234-06 (2006).
5. Bradbury, A. R. M. et al. When monoclonal antibodies are not monospecific: Hybridomas frequently express additional functional variable regions. MAbs 10, 539-546, doi:10.1080/19420862.2018.1445456 (2018).
6. Chen, F. et al. A late Middle Pleistocene Denisovan mandible from the Tibetan Plateau. Nature 569, 409-412, doi:10.1038/s41586-019-1139-x (2019).
7. Hughes, C., Ma, B. & Lajoie, G. A. De novo sequencing methods in proteomics. Methods Mol Biol 604, 105-121, doi:10.1007/978-1-60761-444-9_8 (2010).
8. Jefferis, R. & Lefranc, M. P. Human immunoglobulin allotypes: possible implications for immunogenicity. MAbs 1, 332-338, doi:10.4161/mabs.1.4.9122 (2009).
9. Kaur, H. Characterization of glycosylation in monoclonal antibodies and its importance in therapeutic antibody development. Crit Rev Biotechnol 41, 300-315, doi:10.1080/07388551.2020.1869684 (2021).
10. Jovčevska, I. & Muyldermans, S. The Therapeutic Potential of Nanobodies. BioDrugs 34, 11-26, doi:10.1007/s40259-019-00392-z (2020).
11. Zaroff, S. & Tan, G. Hybridoma technology: the preferred method for monoclonal antibody generation for. Biotechniques 67, 90-92, doi:10.2144/btn-2019-0054 (2019).
12. Alfaro, J. A. et al. The emerging landscape of single-molecule protein sequencing technologies. Nat Methods 18, 604-617, doi:10.1038/s41592-021-01143-1 (2021).
13. Toleikis, L. & Frenzel, A. Cloning single-chain antibody fragments (ScFv) from hyrbidoma cells. Methods Mol Biol 907, 59-71, doi:10.1007/978-1-61779-974-7_3 (2012).
14. Lu, R. M. et al. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci 27, 1, doi:10.1186/s12929-019-0592-z (2020).
15. Boyd, D. et al. Isolation and characterization of a monoclonal antibody containing an extra heavy-light chain Fab arm. MAbs 10, 346-353, doi:10.1080/19420862.2018.1438795 (2018).
16. Harris, C. et al. Identification and characterization of an IgG sequence variant with an 11 kDa heavy chain C-terminal extension using a combination of mass spectrometry and high-throughput sequencing analysis. MAbs 11, 1452-1463, doi:10.1080/19420862.2019.1667740 (2019).
17. Regl, C. et al. Dilute-and-shoot analysis of therapeutic monoclonal antibody variants in fermentation broth: a method capability study. MAbs 11, 569-582, doi:10.1080/19420862.2018.1563034 (2019).
18. Strasser, L. et al. Proteomic Profiling of IgG1 Producing CHO Cells Using LC/LC-SPS-MS. Front Bioeng Biotechnol 9, 569045, doi:10.3389/fbioe.2021.569045 (2021).
19. Thakur, A. et al. Identification, characterization and control of a sequence variant in monoclonal antibody drug product: a case study. Sci Rep 11, 13233, doi:10.1038/s41598-021-92338-1 (2021).
20. Benichou, J., Ben-Hamo, R., Louzoun, Y. & Efroni, S. Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology 135, 183-191, doi:10.1111/j.1365-2567.2011.03527.x (2012).
21. VanDuijn, M. M., Dekker, L. J., van IJcken, W. F. J., Sillevis Smitt, P. A. E. & Luider, T. M. Immune Repertoire after Immunization As Seen by Next-Generation Sequencing and Proteomics. Front Immunol 8, 1286, doi:10.3389/fimmu.2017.01286 (2017).
22. Slatko, B. E., Gardner, A. F. & Ausubel, F. M. Overview of Next-Generation Sequencing Technologies. Curr Protoc Mol Biol 122, e59, doi:10.1002/cpmb.59 (2018).
23. VanDuijn, M. M., Dekker, L. J., van, I. W. F. J., Sillevis Smitt, P. A. E. & Luider, T. M. Immune Repertoire after Immunization As Seen by Next-Generation Sequencing and Proteomics. Front Immunol 8, 1286, doi:10.3389/fimmu.2017.01286 (2017).
24. Kunert, R. & Reinhart, D. Advances in recombinant antibody manufacturing. Appl Microbiol Biotechnol 100, 3451-3461, doi:10.1007/s00253-016-7388-9 (2016).
25. Meyer, L. et al. A simplified workflow for monoclonal antibody sequencing. PLoS One 14, e0218717, doi:10.1371/journal.pone.0218717 (2019).
26. Guthals, A. et al. De Novo MS/MS Sequencing of Native Human Antibodies. J Proteome Res 16, 45-54, doi:10.1021/acs.jproteome.6b00608 (2017).
27. Gilchuk, P. et al. Proteo-Genomic Analysis Identifies Two Major Sites of Vulnerability on Ebolavirus Glycoprotein for Neutralizing Antibodies in Convalescent Human Plasma. Frontiers in Immunology 12, doi:10.3389/fimmu.2021.706757 (2021).
28. Rouet, R., Jackson, K. J. L., Langley, D. B. & Christ, D. Next-Generation Sequencing of Antibody Display Repertoires. Front Immunol 9, 118, doi:10.3389/fimmu.2018.00118 (2018).
29. Chan, C. E., Lim, A. P., MacAry, P. A. & Hanson, B. J. The role of phage display in therapeutic antibody discovery. Int Immunol 26, 649-657, doi:10.1093/intimm/dxu082 (2014).
30. Chen, G. & Sidhu, S. S. Design and generation of synthetic antibody libraries for phage display. Methods Mol Biol 1131, 113-131, doi:10.1007/978-1-62703-992-5_8 (2014).
31. Hammers, C. M. & Stanley, J. R. Antibody phage display: technique and applications. J Invest Dermatol 134, 1-5, doi:10.1038/jid.2013.521 (2014).
32. Dang, H. V. et al. Broadly neutralizing antibody cocktails targeting Nipah virus and Hendra virus fusion glycoproteins. Nat Struct Mol Biol 28, 426-434, doi:10.1038/s41594-021-00584-8 (2021).
33. Bratslavsky, G. T., I. in Developmental Therapeutics Vol. 30 v188 (Annals of Oncology, 2019).
34. Ganesan, R., Grewal, I., S. & Singh, S. MATERIALS AND METHODS FOR MODULATING T CELL MEDIATED IMMUNITY. (2020).
35. in News Medical Life Sciences (2021).
36. Nešić, D. et al. Cryo-Electron Microscopy Structure of the αIIbβ3-Abciximab Complex. Arterioscler Thromb Vasc Biol 40, 624-637, doi:10.1161/ATVBAHA.119.313671 (2020).
37. Dai, D. L. et al. Structural Characterization of Endogenous Tuberous Sclerosis Protein Complex Revealed Potential Polymeric Assembly. Biochemistry 60, 1808-1821, doi:10.1021/acs.biochem.1c00269 (2021).
38. Bernard, M., A.; & Tacahdo, S., D. COMPOSITIONS AND METHODS FOR TREATING INFLAMMATORY DISEASES. (2019).
39. Shibata, H. et al. Comparison of different immunoassay methods to detect human anti-drug antibody using the WHO erythropoietin antibody reference panel for analytes. J Immunol Methods 452, 73-77, doi:10.1016/j.jim.2017.09.009 (2018).
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

