Park, M., Nunez de Villavicencio Diaz, T., Lange, V., Lu, W., Le Bihan, T., Ma, B. Exploring the sheep (Ovis aries) immunoglobulin repertoire by next generation sequencing. Molecular Immunology 156, 20-30 (2023). https://doi.org/10.1016/j.molimm.2023.02.008.
Abstract
Antibody repertoire characterization has often been a challenging feat due to the vastness of B cell receptor (BCR) sequence diversity and the technological limitations to capturing that diversity. The advent of next-generation sequencing (NGS) has since improved the capacity to obtain up to billions of DNA sequences, providing the information required for the comprehensive analyses of antibody repertoires from diverse species (ie. mouse, rabbit, dog, horse, human, etc.).
Though progress has been made in the characterization of diverse immune responses, little is understood about the full immunoglobulin (Ig) repertoire of sheep (Ovis aries), a species widely used for the generation and production of therapeutic antibodies against envenomations and digoxin intoxication. Previous studies have been limited to traditional approaches of cloning transcripts and Sanger sequencing, resulting in the partial characterization of the sheep Ig repertoire.
In this study, we developed an NGS workflow to profile the complete Ig heavy and light chain repertoires in four healthy sheep. Adopting our in-house-developed primers and bioinformatics pipeline, over three million pair-ended sequencing reads were obtained from each of the four sheep, making this study to be the first to achieve this level of sequencing depth in ovine repertoire analysis. We obtained >90% complete antibody sequences and nearly 130,000 unique CDR3 reads for the heavy chain, 48,000 unique CDR3 reads for the kappa chain, and 218,000 unique CDR3 reads for the lambda chain. We also observed biased germline gene usage of variable (V), diversity (D), and joining (J) genes in the heavy and kappa loci only, and immense diversity in the CDR3 sequences.
By conducting an in-depth analysis of non-antigen immunized sheep Ig repertoires via NGS, this data provides further insight into the adaptive immunity of sheep and lays a foundation for future work on immunogenetics and ovine antibody drug development.
Graphical Abstract
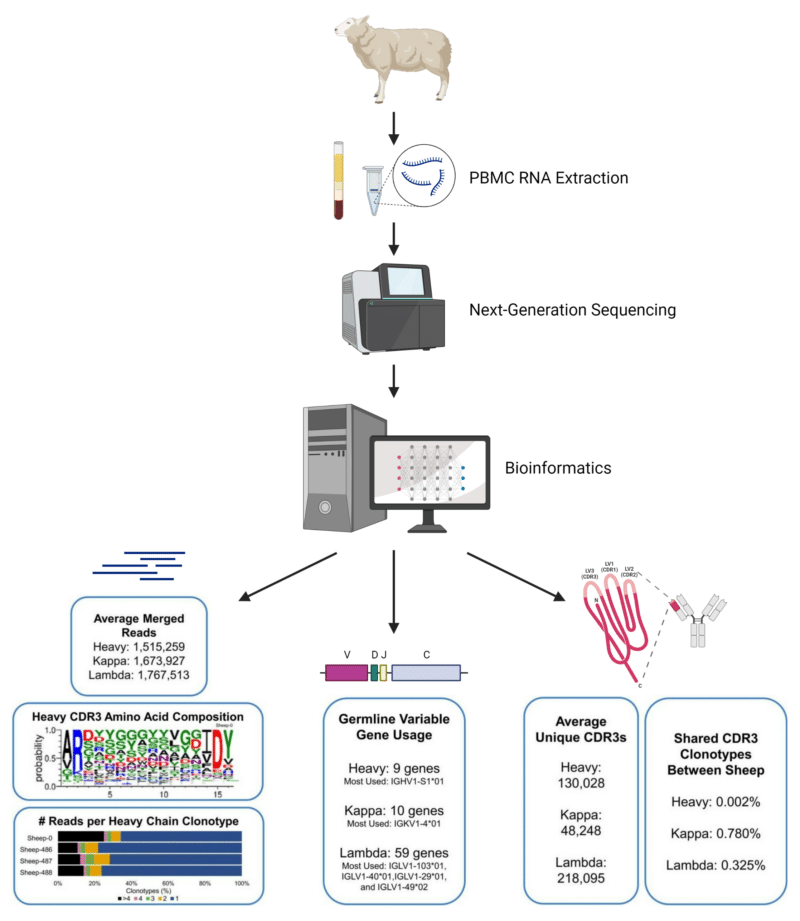
NGS workflow and key results obtained from the comprehensive profiling of the sheep immunoglobulin repertoire.
Key Takeaways
- A workflow for the complete profiling of the sheep immunoglobulin repertoire was developed using next-generation sequencing and bioinformatics methods. Complete antibody sequences (>90%) were obtained and ~130,000, ~48,000, and ~218,000 unique CDR3 reads were obtained for the heavy, kappa, and lambda chains, respectively.
- This comprehensive analysis and insight into the antibody repertoire of healthy sheep can provide the foundation for future immunogenetic studies and anti-ovine antibody drug development.
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics

