Authors:
Teresa Nunez de Villavicencio Diaz, Kyle Suttill, Chelsea Reitzel, Rosalin Dubois,
Qixin Liu, Marko Jović, Dominic Narang, Thierry Le Bihan, Bin Ma
Published: Nov 13, 2023, Revised: Nov 25, 2025
Abstract
In this study, we demonstrate a novel approach for alpaca antibody discovery that bypasses traditional B cell isolation methods. Using mass spectrometry-based de novo protein sequencing (REpAb® platform), we directly sequenced heavy chain-only antibodies (IgG2b/2c) from a commercially available antigen-enriched alpaca IgG preparation raised against rabbit IgG. This protein-first strategy identifies functional antibodies that have already undergone in vivo affinity maturation, thereby eliminating the need for B cell availability and the library-construction biases inherent to conventional phage display approaches.
Ten unique heavy-chain-only antibody sequences were identified and expressed recombinantly, with nine of the 10 demonstrating specific binding to the target antigen. Four antibodies (R1, R4, R7, R9) were selected for comprehensive biophysical characterization using Surface Plasmon Resonance (SPR) for kinetic analysis and epitope binning, and Hydrogen-Deuterium Exchange Mass Spectrometry (HDX-MS) for high-resolution epitope mapping. These techniques revealed diverse binding kinetics and distinct epitopes spanning both the Fab and Fc regions of the antigen, demonstrating the structural diversity of the polyclonal immune response.
This integrated workflow represents the first successful application of direct protein sequencing to alpaca heavy-chain-only antibody discovery, providing a versatile platform for next-generation antibody discovery across species lacking readily available B cell or genomic information.
Key Takeaways
- First report of de novo sequencing of heavy chain-only antibodies (IgG2b/2c) directly from a polyclonal mixture, bypassing B cell isolation and yielding high-affinity binders against a complex antigen (rabbit IgG).
- SPR kinetic analysis and epitope binning revealed diverse binding profiles and epitope competition patterns among four lead antibody candidates.
- HDX-MS epitope mapping identified distinct binding sites across Fab and Fc regions of the antigen, demonstrating the structural diversity of the polyclonal immune response.
- Integrated protein-first approach combining mass spectrometry-based sequencing with biophysical characterization (SPR and HDX-MS) accelerates antibody discovery and lead selection without requiring exhaustive candidate production.
Introduction
Camelid Antibodies: A Unique Immune Arsenal
Alpacas and other camelids produce a distinctive antibody repertoire that includes both conventional immunoglobulins and a unique class of heavy-chain-only antibodies. Alongside canonical IgG (IgG1a/b) with the traditional heavy and light chain structure, camelids naturally produce non-canonical IgG subtypes (IgG2b/2c) that consist solely of heavy chains lacking the CH1 domain and any associated light chains (Figure 1). The variable domains of these heavy-chain-only antibodies, termed VHH domains, represent the smallest functional antigen-binding units (~15 kDa) and can be expressed recombinantly as isolated single-domain antibodies, also known as “nanobodies.” These VHH-derived nanobodies offer significant advantages for therapeutic and diagnostic applications. One of their key advantages is high stability, small size that enables access to cryptic epitopes, ease of production, and robust tissue penetration. Additionally, their modular architecture facilitates engineering of bispecific and multispecific constructs, enabling simultaneous targeting of multiple antigens or epitopes, a capability increasingly valuable for complex therapeutic applications such as tumour-targeted drug delivery and immune cell engagement.

Figure 1. Structural comparison of canonical and non-canonical alpaca IgG antibodies. Alpacas produce both canonical IgG (IgG1a/b, left) with standard heavy and light chain pairing (VH-CH1-CH2-CH3 and VL-CL), and non-canonical heavy chain-only IgG (IgG2b/2c, middle) lacking light chains and the CH1 domain (VHH-CH2-CH3). Nanobodies (right) are recombinant constructs consisting of isolated VHH domains and do not occur naturally. Hinge region sequences distinguish IgG subtypes, with conserved cysteines (red) forming interchain disulphide bonds.
Limitations of Conventional VHH Discovery
Traditional nanobody discovery workflows rely on B cell isolation following immunization. The process involves extracting peripheral blood mononuclear cells (PBMCs), constructing VHH libraries through PCR amplification, displaying these libraries (typically in phage or yeast), and selecting binders through multiple rounds of panning against the target antigen. Selected candidates are then expressed, usually in E. coli, for downstream characterization. While this approach has yielded numerous successful nanobodies, it faces several inherent limitations. The method is fundamentally constrained by the availability of B cells in peripheral blood at the time of sampling, which may not accurately reflect the full diversity of the immune response. Library construction introduces bias during PCR amplification and cloning, potentially losing rare but high-value sequences. Critically, selection occurs at the DNA level via display technologies, meaning the actual binding assessment occurs on displayed constructs rather than on the mature, functional antibodies that circulate in the animal. Consequently, many selected candidates fail to express properly or lack the binding characteristics observed during selection, requiring screening of large numbers of clones to identify functional leads.
Direct Protein Sequencing, Accessing Proven Binders
Our proposed approach fundamentally differs by directly sequencing heavy-chain-only IgG antibodies (IgG2b/2c) from antigen-enriched preparations. This protein-first strategy offers distinct advantages over conventional methods. Most importantly, we sequence only antibodies that have already demonstrated functionality in the animal—these are proteins that successfully bind antigen, undergo in vivo affinity maturation through multiple rounds of somatic hypermutation, and achieve high-affinity binding in their natural physiological context. By performing stringent antigen-based enrichment of functional protein antibodies before sequencing, we directly access the most biologically relevant binders without depending on B cell availability in circulation or on potential library construction biases. The antibodies we sequence have already demonstrated functionality in vivo. They are mature, affinity-matured proteins selected through the animal’s own immune system, rather than candidates that may or may not function when recombinantly expressed and folded.
Study Overview
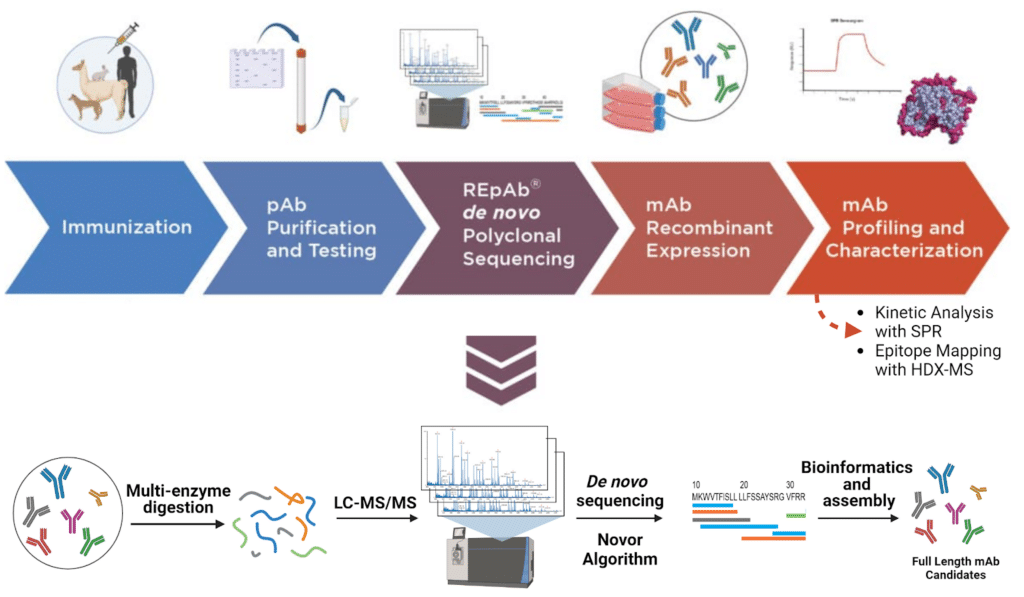
This study demonstrates the power of a protein-based discovery approach using a commercially available antigen-enriched alpaca IgG preparation. An alpaca was immunized against rabbit IgG, yielding a polyclonal mixture containing both canonical and non-canonical alpaca IgG antibodies. We sequenced the heavy chain-only antibodies (IgG2b/2c) directly from this preparation. Following multi-protease digestion, mass spectrometry-based de novo peptide sequencing was performed, and full-length antibody sequences were assembled using machine learning-based bioinformatics integrated into the REpAb® platform (Figure 2). Ten unique, full-length, heavy-chain-only antibody sequences were discovered and expressed recombinantly. We then performed a comprehensive characterization of selected candidates using ELISA for initial binding confirmation, Surface Plasmon Resonance (SPR) for detailed kinetic analysis and epitope binning, and Hydrogen-Deuterium Exchange Mass Spectrometry (HDX-MS) for high-resolution epitope mapping. This integrated workflow demonstrates how direct protein sequencing enables the rapid discovery of functional, high-affinity antibodies, coupled with detailed biophysical characterization, to support the selection and development of leads.
Figure 2. Antibody discovery and characterization pipeline
Methods
De Novo Sequencing
An antigen-enriched alpaca IgG preparation obtained from a commercial source containing both canonical and non-canonical IgGs, confirmed by differential migration on SDS-PAGE gel, was digested with several different protease enzymes. The digests were analyzed by liquid chromatography-tandem mass spectrometry using an Orbitrap Eclipse Series Instrument (Thermo Fisher Scientific, CA, US) coupled to an Evosep One LC system (Evosep, Denmark). De novo peptide sequencing was performed using proprietary software that is an evolution of the Novor search algorithm. Full-length antibody sequences were assembled using the REpAb® platform, which incorporates machine learning-based bioinformatics, as described in Le Bihan et al. (2024) (doi.org/10.1038/s41467-024-53105-8). Ten unique heavy-chain-only antibody sequences were identified and expressed recombinantly as human Fc fusions via Sino Biological. ELISA confirmed binding affinity.
Epitope Mapping with Hydrogen-Deuterium Exchange Mass Spectrometry (HDX-MS)
The HDX-MS experiments were carried out using the Trajan automation platform and Waters Cyclic IMS MS. The antigen digestion was performed using a pepsin/protease XIII column, and peptides were identified using the Waters™ SELECT SERIES Cyclic IMS mass spectrometer. For D2O labelling experiments, two different time points were used (10 and 60 minutes) in both the free and bound states. PLGS was used to prepare a peptide library, and DynamX 3.0 was used for deuterium uptake analysis.
Kinetic Analysis and Epitope Binning with Surface Plasmon Resonance (SPR)
Using the OpenSPR instrument (Nicoya Lifesciences, Kitchener, ON). Each ligand used for multicycle kinetic analysis and epitope binning was immobilized in the active channel at 20 μg/ml using a flow rate of 10 μl/min. Amine coupling was performed using the standard EDC/NHS chemistry on High-Capacity Carboxyl Sensors. Blocking of the active and reference channels was performed by passing BSA at a concentration of 260 μg/ml following the ligand immobilization in the active channel. Finally, 1 M Tris-based solution was used for reaction quenching. The optimized immobilization setup provided sufficient immobilization levels, well above the minimum requirement of the OpenSPR system (Rmax [P17] = 236.2 RU and Rmax [CD3ε] = 272 RU).
For each multicycle kinetic analysis run, analytes were prepared in the running buffer PBSTE (0.1% Tween, 10 mM EDTA) at the highest concentration to be injected and further diluted in a 3-fold series to the lowest concentration to be injected. The analyte was then injected following buffer blank(s), in the order of low to high. Data was exported into TraceDrawer analysis software for kinetic fitting using a 1:1 binding model.
Results and Discussion
10 Unique Heavy Chain-Only Antibody Sequences Identified from the Polyclonal Mixture
A commercial antigen-enriched alpaca IgG preparation raised against rabbit IgG was obtained as starting material for this proof-of-concept study. Non-reducing SDS-PAGE analysis confirmed the presence of both canonical IgG (IgG1a/b, ~150 kDa) and heavy chain-only non-canonical IgG (IgG2b/2c, ~90-100 kDa) in the preparation (Figure 3A). This mixture, containing both antibody formats, presented an added challenge for selective sequencing and validated the capability of the REpAb® platform to distinguish and identify heavy chain-only antibodies from complex polyclonal preparations. We performed multi-protease digestion followed by LC-MS/MS analysis to sequence the antigen-enriched alpaca IgG preparation. De novo peptide sequencing identified multiple unique CDR3 sequences, which were then assembled into full-length heavy-chain-only antibody sequences through contig elongation using the REpAb® platform. Non-canonical IgG2b and IgG2c sequences were distinguished by their characteristic hinge region sequences (Figure 3B). The ten most abundant sequences, ranked by CDR3 peptide intensity, coverage, and sequence quality scores, were selected for recombinant expression. All ten heavy chain-only antibody sequences were expressed as VHH-human Fc fusion proteins to facilitate downstream characterization. Additionally, a subset was recombinantly expressed as isolated VHH constructs. ELISA analysis confirmed that 9 of 10 recombinant antibodies bound specifically to the rabbit IgG antigen, validating the functional relevance of the sequences identified directly from the polyclonal mixture (Figure 3C).
Figure 3. De novo sequencing and recombinant antibody binding validation. Figure 3A. Non-reducing SDS-PAGE gel demonstrating the presence of both canonical IgG1a/b and heavy chain-only IgG2b/2c in the antigen-enriched alpaca IgG preparation. Figure 3B. De novo sequencing assembly of a representative heavy chain-only antibody (IgG2c) from the polyclonal mixture. The top line displays region assignments according to the CHOTHIA numbering scheme: green represents framework regions, orange denotes CDR3, and blue indicates the constant region (CH2/CH3). The consensus sequence is shown in the middle line. The underlined sequence AHHSEDPSSKCPKCP is the hinge region specific to IgG2c, which distinguishes it from IgG2b. Colored sequences below represent peptides from different protease digests: chymotrypsin (green), trypsin (blue), lysC (light orange), AspN (dark orange), and pepsin (black). More than 30 peptide observations support each amino acid position; only the top 13 most intense peptides per region are shown. Figure 3C. ELISA binding analysis of ten recombinant alpaca heavy chain-only antibodies expressed as VHH-human Fc fusions. Nine of ten antibodies (R1-R9) demonstrated specific binding to rabbit IgG antigen with apparent affinities in the low nanomolar range. Human IgG (hIgG) served as a negative control.
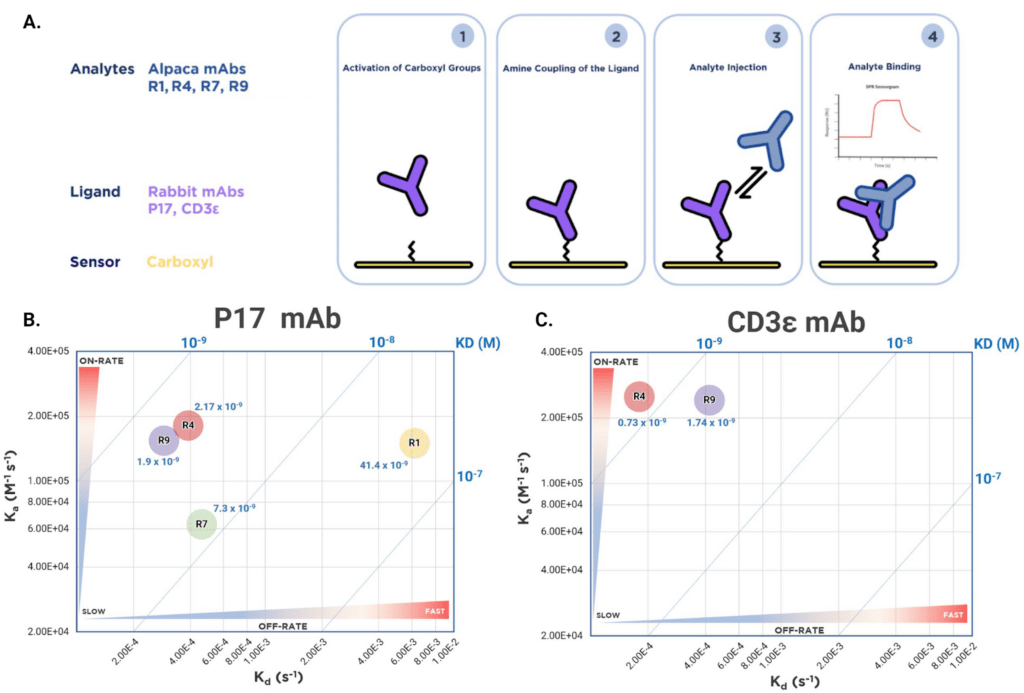
Kinetic Analysis with SPR Reveals Four High Affinity Binders
Of the ten heavy chain-only antibody sequences identified, four (R1, R4, R7, and R9) were selected for detailed kinetic characterization. Using SPR, full kinetic profiles (kon, koff, and Kd) were determined for each antibody against two rabbit monoclonal antigens, a kappa light chain-containing anti-beta integrin IgG (P17, internal designation) and a lambda light chain-containing anti-CD3ε IgG (Figure 4A). Each of the four binders exhibited a unique binding profile beyond what was observed in ELISA. Key distinguishing characteristics include the slow on-rate of R7 (6.3×105 M-1 s-1) and the fast off-rate of the R1 clone (6.19×10-5 s-1) (Figure 4B). R4 and R9 exhibited similar kinetic profiles with high affinity toward both anti-CD3ε and anti-beta integrin antigens, but showed a slight preference for anti-CD3ε (Figure 4C). In contrast, R1 and R7 exhibited specific binding in the nanomolar range exclusively towards the kappa light chain-containing IgG, P17 (Figure 4B).
The lower affinity of R1 is primarily driven by its off-rate, which is an order of magnitude faster than those of R4, R7, and R9. The weaker affinity of R7 for P17 is primarily due to its slower on-rate compared to those of the other antibodies.
Figure 4. Kinetic analysis and iso-affinity profiling of four recombinant alpaca heavy chain-only antibodies. Figure 4A. SPR experimental workflow for kinetic analysis. Rabbit monoclonal antibodies (P17, anti-CD3ε) were immobilized on a carboxyl sensor chip. Alpaca antibodies (R1, R4, R7, R9) were injected as analytes for kinetic measurements. Figure 4B. Iso-affinity kinetic plot for alpaca antibodies binding to P17. Two-dimensional plot of association rate constant (Ka) versus dissociation rate constant (Kd). Blue diagonal lines represent equilibrium dissociation constants (Kd) calculated from the ratio of rate constants. Regions labelled indicate kinetic behaviour: slow/fast on-rate and slow/fast off-rate. Figure 4C. Iso-affinity kinetic plot for alpaca antibodies binding to anti-CD3ε, displayed as in Figure 4B, highlighting difference in binding specificity of nanobodies towards the kappa light chain-containing P17 IgG and the lambda light chain-containing CD3ε IgG.
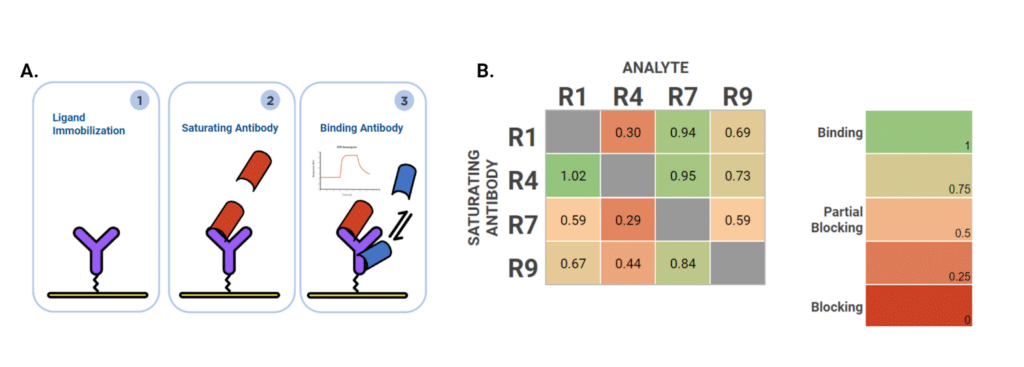
Epitope Binning of Four Alpaca mAbs on P17 Antigen
To investigate epitope diversity using SPR, we conducted a 4×4 tandem epitope binning experiment to examine the epitope distribution of four alpaca heavy chain-only antibodies against the P17 rabbit monoclonal antigen. P17 was immobilized on the sensor surface, followed by sequential injections of a saturating antibody (blocker) and a binding antibody (analyte) (Figure 5A). Tandem epitope binning was performed in both directions, with each antibody serving as both the saturating and binding antibody. Saturation efficiency was assessed using the same antibody for both blocking and binding (diagonal values in Figure 5B).
When used as a saturating antibody, R4 showed the highest compatibility, with the other three antibodies exhibiting minimal reduction in binding response (0-27% blocking, indicating non-overlapping or partially overlapping epitopes). Conversely, blocking with R7 dramatically reduced epitope accessibility for the other antibodies (41-71% blocking, Figure 5B), suggesting that R7 binds to a central or sterically hindering epitope. To further elucidate antibody-antigen interactions at the structural level, HDX-MS analysis was performed.
Figure 5. Epitope binning analysis of four alpaca heavy chain-only antibodies against P17. Figure 5A. SPR experimental workflow for tandem epitope binning. P17 antigen was immobilized on the sensor surface. A saturating antibody was first injected, followed by the injection of a binding antibody (analyte). The binding of the analyte indicates non-competing epitopes, while blocking indicates overlapping or competing epitopes. Figure 5B. Epitope binning heatmap showing fractional binding of analyte antibodies in the presence of saturating antibodies. Each antibody (R1, R4, R7, R9) was tested at 100 nM as both the saturating antibody (y-axis) and analyte (x-axis) in a 4×4 matrix. Values represent the fraction of analyte binding relative to binding on unblocked antigen (1.0 = no blocking/non-competing epitopes; 0 = complete blocking/competing epitopes). Color scale: green = binding (non-competing), yellow/orange = partial blocking (partially overlapping), red = blocking (competing epitopes).
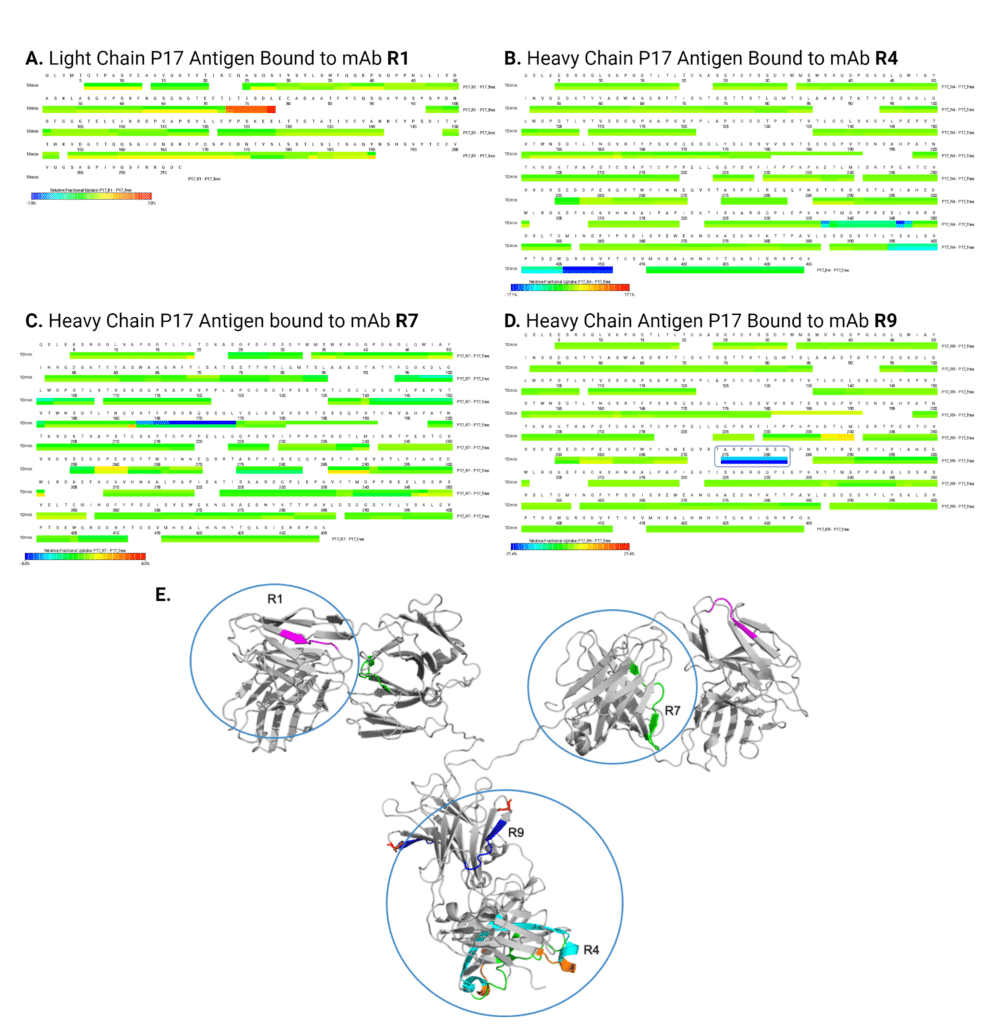
Epitope Mapping with HDX-MS Identified Four Unique Binding Epitopes on the Antigen
Using HDX-MS, we mapped the epitopes of the four recombinant alpaca heavy chain-only antibodies (R1, R4, R7, and R9) on the rabbit P17 IgG antigen. Each antibody exhibited a distinct binding footprint on the antigen, as evidenced by changes in relative fractional uptake between the antibody-bound and free states of the antigen (Figure 6A-D).
Notably, R1 binding resulted in increased deuterium uptake in certain regions of the light chain (indicated by red in the heatmap, Figure 6A), suggesting conformational changes or increased solvent accessibility upon antibody binding. This is unusual compared to the other three antibodies (R4, R7, R9), which showed primarily decreased deuterium uptake (blue regions, Figure 6B-D), indicating protection of the antigen backbone from solvent exchange upon antibody binding—a typical signature of epitope engagement.
HDX-MS epitope mapping revealed that R1 and R7 bind to the Fab region of the P17 antigen, whereas R4 and R9 bind to the Fc region (Figure 6E). This epitope diversity demonstrates that the polyclonal alpaca immune response generated antibodies targeting distinct structural regions of the antigen, consistent with the epitope binning results obtained by SPR.
Figure 6. Epitope mapping of four alpaca heavy chain-only antibodies by HDX-MS. Figure 6A-D. Deuterium uptake heatmaps showing peptide-level protection patterns upon antibody binding to P17 antigen. Each panel shows a heatmap of differential deuterium uptake comparing antibody-bound versus free states: Figure 6A. Light chain (LC) heatmap upon R1 binding, Figure 6B. Heavy chain (HC) heatmap upon R4 binding, Figure 6C. HC heatmap upon R7 binding, and Figure 6D. HC heatmap upon R9 binding. Color scale represents relative deuterium uptake: blue = decreased uptake (protection/binding interface), green = no change, red = increased uptake (conformational change or increased solvent accessibility). Figure 6E. Structural mapping of epitopes on a homology model of rabbit IgG (gray, generated in PyMOL). Epitopes for R1 (magenta), R4 (cyan), R7 (green), and R9 (blue) are highlighted on the structure, demonstrating binding diversity across both Fab and Fc regions of the antigen. R1 and R7 target the Fab region, while R4 and R9 target the Fc region.
Conclusions
To our knowledge, this study represents the first successful application of de novo protein sequencing to discover and characterize alpaca heavy-chain-only antibodies directly from an antigen-enriched IgG preparation, thereby bypassing the need for B cell isolation entirely (November 2023). By sequencing functional antibodies that have already undergone in vivo affinity maturation, this approach identifies high-affinity binders without being limited by the availability of B cells in peripheral circulation. Furthermore, we demonstrate an integrated workflow combining de novo polyclonal antibody sequencing with biophysical characterization techniques (SPR and HDX-MS) to enable comprehensive lead identification and selection.
Alpacas produce a unique subclass of heavy chain-only antibodies (IgG2b/2c) that lack both light chains and the CH1 domain. These structurally distinct immunoglobulins offer significant potential for biologics development and engineering, as their variable domains (VHH) can be expressed as small (~15 kDa), highly stable nanobodies with exceptional tissue penetration and access to cryptic epitopes. The REpAb® platform’s ability to sequence these antibodies directly at the protein level demonstrates its adaptability to diverse species and antibody formats, particularly those lacking readily available B cell or genomic information. This protein-first methodology is broadly applicable to exploring novel antibody architectures, showcasing its versatility for next-generation antibody discovery across multiple therapeutic modalities.
Disclaimer: This document was drafted by the authors, refined with assistance from Claude.ai (Anthropic), and finalized through human review and editing.
More Information on Antibody Discovery and Characterization
Antibody Discovery Services
Mass spectrometry based in vivo antibody discovery platform, that sequences functional antibodies directly from serum or purified protein samples.
Explore Antibody Discovery Services
SPR Kinetic Analysis and Epitope Binning
Our Surface Plasmon Resonance (SPR) service can determine the on-rate (Kon), off-rate (Koff), dissociation constant (Kd), and affinity (KD) for antibody-antigen interaction.
Explore SPR Services
HDX-MS Epitope Mapping
Mass spectrometry based epitope mapping service, that can identify the binding site of an antibody to its corresponding antigen with the highest confidence and resolution.
Explore Epitope Mapping Services
Talk to Our Scientists.
We Have Sequenced 10,000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and developed the first recombinant polyclonal antibody diagnostics.
Talk to Our Scientists.
We Have Sequenced 9000+ Antibodies and We Are Eager to Help You.
Through next generation protein sequencing, Rapid Novor enables timely and reliable discovery and development of novel reagents, diagnostics, and therapeutics. Thanks to our Next Generation Protein Sequencing and antibody discovery services, researchers have furthered thousands of projects, patented antibody therapeutics, and ran the first recombinant polyclonal antibody diagnostics